
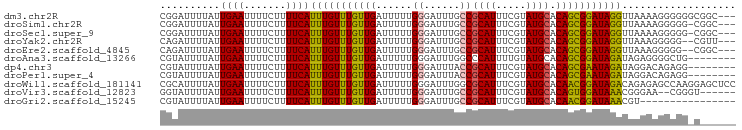
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,856,744 – 16,856,837 |
| Length | 93 |
| Max. P | 0.648201 |

| Location | 16,856,744 – 16,856,837 |
|---|---|
| Length | 93 |
| Sequences | 11 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.97 |
| Shannon entropy | 0.31896 |
| G+C content | 0.37383 |
| Mean single sequence MFE | -20.43 |
| Consensus MFE | -14.17 |
| Energy contribution | -13.67 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.648201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
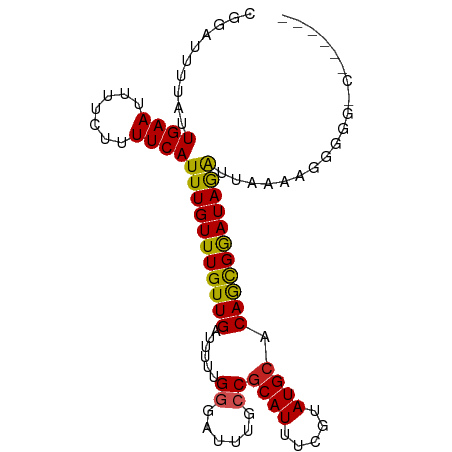
>dm3.chr2R 16856744 93 + 21146708 CGGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGGUUAAAAGGGGGGCGGC--- ..............((..(((((.((((((((((((......((......))((((.....)))).)))))))))))).)))))..)).....--- ( -22.80, z-score = -1.73, R) >droSim1.chr2R 15511165 92 + 19596830 CGGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGGUUAAAAGGGGG-CGGC--- ..............((..(((((.((((((((((((......((......))((((.....)))).)))))))))))).)))))..))-....--- ( -22.50, z-score = -1.70, R) >droSec1.super_9 187687 92 + 3197100 CGGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGGUUAAAAGGGGG-CGGC--- ..............((..(((((.((((((((((((......((......))((((.....)))).)))))))))))).)))))..))-....--- ( -22.50, z-score = -1.70, R) >droYak2.chr2R 8798852 91 - 21139217 CAGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGGUUAAAGGGGG--CGUU--- ...............(..(((((.((((((((((((......((......))((((.....)))).)))))))))))).)))))..)--....--- ( -21.60, z-score = -1.79, R) >droEre2.scaffold_4845 11000121 91 + 22589142 CAGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGGUUAAAGGGGG--CGGC--- ...............(..(((((.((((((((((((......((......))((((.....)))).)))))))))))).)))))..)--....--- ( -21.60, z-score = -1.60, R) >droAna3.scaffold_13266 16158084 88 - 19884421 CGUAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGGCCCAUUUUGUAUGCACAGCGGAUAGAUAGAGGGGCUG-------- ..................((((((((((((((((((.....((((......))))...((....)))))))))))))).))))))...-------- ( -19.50, z-score = -1.32, R) >dp4.chr3 11487184 88 - 19779522 CGUAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUACCGCAUUUCGUAUGCACAGCGAAUAGAUAGGACAGAGG-------- ................((((.(((((((((((((((......((......))((((.....)))).)))))))))))).))).)))).-------- ( -22.00, z-score = -3.29, R) >droPer1.super_4 4988168 88 - 7162766 CGUAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUACCGCAUUUCGUAUGCACAGCGAAUAGAUAGGACAGAGG-------- ................((((.(((((((((((((((......((......))((((.....)))).)))))))))))).))).)))).-------- ( -22.00, z-score = -3.29, R) >droWil1.scaffold_181141 5194820 96 + 5303230 CGCAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGGCGCAUUUCGUAUGCACAACGGAUAGACAGAGAGCCAAGGAGCUCC ..........((((.......))))...((((.(((.((((((..(((((..((((.....))))....)))))...))))))..))).))))... ( -17.80, z-score = 0.46, R) >droVir3.scaffold_12823 683043 88 - 2474545 GGUAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGUGGAUAAACGGGAA--CGGGU------ ................(((((..(.((((((..(((......((......))((((.....)))).)))..)))))).)..))--.))).------ ( -16.40, z-score = -0.25, R) >droGri2.scaffold_15245 16248247 80 + 18325388 CGUAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAACGGAUAAACGU---------------- (((.......((((.......)))).((((((((((......((......))((((.....)))).))))))))))))).---------------- ( -16.00, z-score = -1.45, R) >consensus CGGAUUUUAUUGAAUUUUCUUUUCAUUUGUUUGUUGAUUUUUGGGAUUUGCCGCAUUUCGUAUGCACAGCGGAUAGAUUAAAAGGGGG_C______ ..........((((.......))))(((((((((((......((......))((((.....)))).)))))))))))................... (-14.17 = -13.67 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:42 2011