| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,841,217 – 16,841,311 |
| Length | 94 |
| Max. P | 0.614346 |

| Location | 16,841,217 – 16,841,311 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 66.18 |
| Shannon entropy | 0.71200 |
| G+C content | 0.45998 |
| Mean single sequence MFE | -20.30 |
| Consensus MFE | -10.87 |
| Energy contribution | -10.87 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.69 |
| Mean z-score | -0.25 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16841217 94 - 21146708 --CCGCGCUGCAGGAUAUCGCAUUG-----UGUGCGCGAUGGCCUCAAAUUUAAUAAUAAAAACGCCGUCAAGUUCCUGUUCGUUCGAUUCUGUCUAGAUU------- --..(((..((((((...(((((..-----.))))).((((((.....................))))))....)))))).))).................------- ( -21.90, z-score = -0.27, R) >droSim1.chr2R 15495450 94 - 19596830 --CCUCGCUGCAGGAUAUCGCAUUG-----UGUGCGCGAUGGCCUCAAAUUUAAUAAUAAAAACGCCGUCAAGUUCCUGUUCG------UUCGAUUCUGUCUAGAUU- --..(((..((((((...(((((..-----.))))).((((((.....................))))))....))))))...------..))).............- ( -20.50, z-score = -0.43, R) >droSec1.super_9 172203 94 - 3197100 --CCUCGCUGCAGGAUAUCGCAUUG-----UGUGCGCGAUGGCCUCAAAUUUAAUAAUAAAAACGCCGUCAAGUUCCUGUUCG------UUCGAUUCUGUCUAGAUU- --..(((..((((((...(((((..-----.))))).((((((.....................))))))....))))))...------..))).............- ( -20.50, z-score = -0.43, R) >droYak2.chr2R 8782788 96 + 21139217 --CCUCGCUGCAGGAUAUCGCAUUG-----UGUGCGCGAUGGCCUCAAAUUUAAUAAU-AAAACGCCGUCAAGUUCCUGUUCCAGUUCGAUUCUCUCUGAGAUU---- --.((((..((((((...(((((..-----.))))).((((((.....(((....)))-.....))))))....))))))..).....((.....)).)))...---- ( -22.20, z-score = -0.76, R) >droEre2.scaffold_4845 10984422 99 - 22589142 --CCUCGCUGCAGGAUAUCGCAUUG-----UGUGCGCGAUGGCCUCAAAUUUAAUAAU-AAAACGCCGUCAAGUUCCUGUUCAUCCUGUUUCGGUUCUGUCUAGAUU- --..(((..(((((((..(((((..-----.))))).((((((.....(((....)))-.....))))))............)))))))..))).............- ( -23.90, z-score = -1.33, R) >droAna3.scaffold_13266 16141462 103 + 19884421 --GCUCUCGGCAGGAUAUCGUGUUGUAUG-UAUGUGCAAUGGCCUCAAAUUUAAUAAU-AAAUCGCCGUCAAGUUCCUGUU-CAGCCACUCGCUCGCUCAGUCAGUCA --(((...(((((((......(((((((.-...)))))))(((.....(((((....)-)))).))).......)))))))-.))).(((.(((.....))).))).. ( -19.80, z-score = 0.10, R) >dp4.chr3 11471000 88 + 19779522 --GCUCGCGGCAGGAUGUCGCAUUG-----UGUGUGCAAUGGCCUCAAAUUUAAUAAU-AAAACGCCGUCAAGUUCCUGCUCAGGCUGCUCUGUCU------------ --((..(.(((((((....((((..-----...)))).(((((.....(((....)))-.....))))).....))))))))..))..........------------ ( -22.00, z-score = 0.08, R) >droPer1.super_4 4971929 88 + 7162766 --GCUCGCGGCAGGAUGUCGCAUUG-----UGUGUGCAAUGGCCUCAAAUUUAAUAAU-AAAACGCCGUCAAGUUCCUGCUCAGGCUGCUCUGUCU------------ --((..(.(((((((....((((..-----...)))).(((((.....(((....)))-.....))))).....))))))))..))..........------------ ( -22.00, z-score = 0.08, R) >droWil1.scaffold_181141 5173797 90 - 5303230 CACCGCUCGGCAGGAUAUCGUUCUAUGCAUUGUGAGCAAGGACCUCAAAUUUAAUAAA-AAA-CGCCGUCAAGUUCCUGUUACGUUCACUAU---------------- ........(((((((....(((((.(((.......))))))))...............-..(-(........)))))))))...........---------------- ( -14.20, z-score = 0.70, R) >droMoj3.scaffold_6496 25763242 75 + 26866924 --UGGCGCCACAGGGCGUCGCAUUG-----UGUGCGAAUUUUAAUAAAAUUCGACAAGUUUUUCUCCUUUCAGGUCGAGUCU-------------------------- --.((((((....))))))((((..-----.)))).............(((((((..(............)..)))))))..-------------------------- ( -19.10, z-score = -1.03, R) >droGri2.scaffold_15245 16231315 88 - 18325388 --GGGCGCCACAGGACAUCGCUUUUUG---UGUGUGCAACGGCCUUAAAUUUAAUA---AAAUCGCCGUCAAGUUUGUGCUUCUGAUUCAUAUUCG------------ --..(((((((((((.......)))))---)).)))).(((((.............---.....))))).((((....))))..............------------ ( -17.17, z-score = 0.59, R) >consensus __CCUCGCUGCAGGAUAUCGCAUUG_____UGUGCGCAAUGGCCUCAAAUUUAAUAAU_AAAACGCCGUCAAGUUCCUGUUCC_GCU_CUUUGUCU____________ .........((((((...(((((........)))))..(((((.....................))))).....))))))............................ (-10.87 = -10.87 + 0.01)

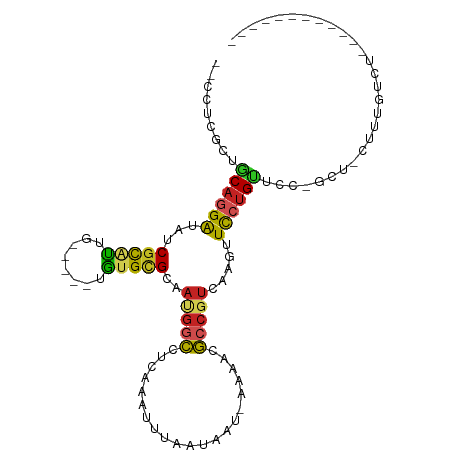

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:40 2011