| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,482,351 – 3,482,446 |
| Length | 95 |
| Max. P | 0.914308 |

| Location | 3,482,351 – 3,482,446 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.81 |
| Shannon entropy | 0.44173 |
| G+C content | 0.57142 |
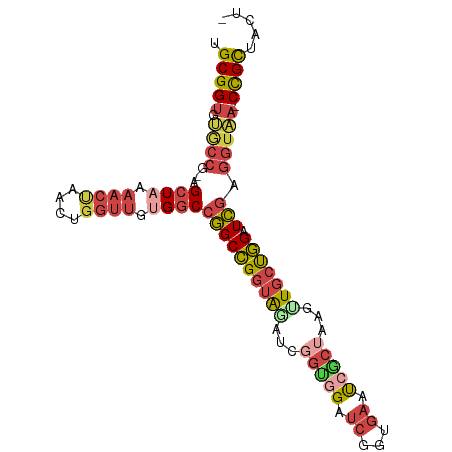
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -21.92 |
| Energy contribution | -23.85 |
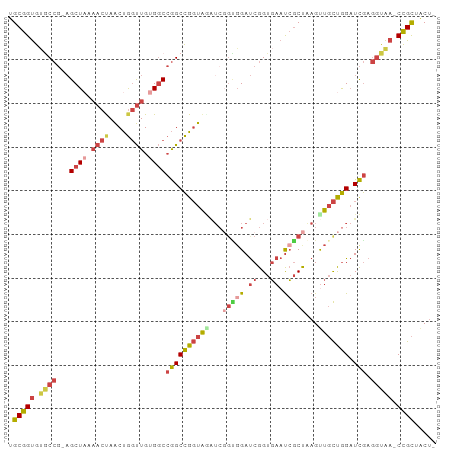
| Covariance contribution | 1.93 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914308 |
| Prediction | RNA |
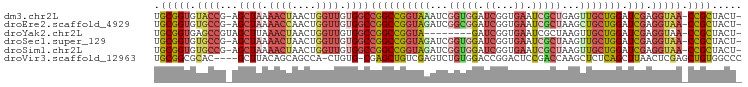
Download alignment: ClustalW | MAF
>dm3.chr2L 3482351 95 + 23011544 UGCGGUGUACCG-AGCUAAAACUAACUGGUUGUGGCCGGCCGGUAAAUCGGUGGAUCGGUGAAUCGCUGAGUUGCUGGAUCGAGGUAA-CCGCUACU- .(((((.((((.-.((((.((((....)))).))))((((((((((.(((((((.((...)).))))))).))))))).))).)))))-))))....- ( -39.40, z-score = -3.33, R) >droEre2.scaffold_4929 3517091 95 + 26641161 UGCGGUGUGCCG-AGCUAAAACCAACUGGUUGUGGCCGGCCGGUAGAUCGGCGGAUCGGUGAAUCGCUAAGCUGCUGGAUCGAGGUAA-CCGCUACU- .(((((.((((.-.((((.((((....)))).))))((((((((((...(((((.((...)).)))))...))))))).))).)))))-))))....- ( -41.30, z-score = -2.65, R) >droYak2.chr2L 3469920 88 + 22324452 UGCGGUGAGCCGUAGCUUAAACUAACUGGUUGUGGCCGGCCGGUA--------GAUCGGUGAAUCGCUAAGUUGCUGGAUCGAGGUAA-CCGCUACU- .(((((..(((...(((..((((....))))..)))(((((((((--------(...((((...))))...))))))).))).))).)-))))....- ( -29.90, z-score = -0.73, R) >droSec1.super_129 44331 95 + 51580 UGCGGUGUGCCG-AGCUAAAACUAACUGGUUGUGGCCGGCCGGUAGAUCGGUGGAUCGGUGAAUCGCUAAGUUGCUGGAUCGAGGUAA-CCGCUACU- .(((((.((((.-.((((.((((....)))).))))((((((((((...(((((.((...)).)))))...))))))).))).)))))-))))....- ( -34.70, z-score = -1.43, R) >droSim1.chr2L 3435707 95 + 22036055 UGCGGUGUGCCG-AGCUAAAACUAACUGGUUGUGGCCGGCCGGUAGAUCGGUGGAUCGGUGAAUCGCUAAGUUGCUGGAUCGAGGUAA-CCGCUACU- .(((((.((((.-.((((.((((....)))).))))((((((((((...(((((.((...)).)))))...))))))).))).)))))-))))....- ( -34.70, z-score = -1.43, R) >droVir3.scaffold_12963 4888402 92 - 20206255 UGCGGCGCAC----GCUUACAGCAGCCA-CUGUG-CGAGCUGUCGAGUCUGUGGACCGGACUCCGACCAAGCUCUCAGCUUAACUCGAGCUGUGGCCC ...(((((((----(((......)))..-..)))-)(((((((((((((((.....)))))).))))..))))).((((((.....))))))..))). ( -36.60, z-score = -0.92, R) >consensus UGCGGUGUGCCG_AGCUAAAACUAACUGGUUGUGGCCGGCCGGUAGAUCGGUGGAUCGGUGAAUCGCUAAGUUGCUGGAUCGAGGUAA_CCGCUACU_ .((((..((((...((((.((((....)))).))))((((((((((...(((((.((...)).)))))...))))))).))).))))..))))..... (-21.92 = -23.85 + 1.93)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:59 2011