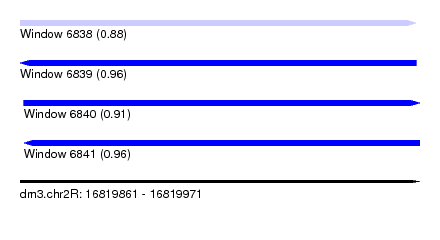
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,819,861 – 16,819,971 |
| Length | 110 |
| Max. P | 0.964671 |

| Location | 16,819,861 – 16,819,970 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.48076 |
| G+C content | 0.38826 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.879980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16819861 109 + 21146708 AUUUUGCUGCCAUGCAAUUAGUUCGCCUAUCGCCGU-------ACGAAA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA .......((.(((((((.......(((.((((..((-------((....----...............))))...)))))))........(((((((.....)))))))))))))).)). ( -27.21, z-score = -0.97, R) >droPer1.super_4 4947497 108 - 7162766 UUUUUGUUUCUAUGCAAUUAACUCGCUUAUCGGCAA--------CGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGUAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACAA ...(((((...((((((((.(((((....(((....--------)))..----.)))))..((((...(((.((........))))).))))(((((.....)))))))))))))))))) ( -26.80, z-score = -1.69, R) >dp4.chr3 11446372 108 - 19779522 UUUUUGUUUCUAUGCAAUUAACUCGCUUAUCGGCAA--------CGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGCAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACAA ...(((((...((((((((.(((((....(((....--------)))..----.)))))..((((....(((........))).....))))(((((.....)))))))))))))))))) ( -27.00, z-score = -1.63, R) >droAna3.scaffold_13266 16118502 120 - 19884421 UUUUUGCUUCCAUGCAAUUAACUCGCCUAUCGCCUAUCGCCCAACGAGACCCAACCGGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAAUUGAUUGAAUUGCAUGACAA ..........(((((((..((((((.((((((.....(((.....(((.((.....))))).......)))....))))))...))))))(((((((.....)))))))))))))).... ( -31.30, z-score = -2.82, R) >droEre2.scaffold_4845 10961928 109 + 22589142 AUUUUGCUGCCAUGCAAUUAGUUCGCCUAACGCCGU-------ACGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA .......((.(((((((..............(((((-------.((((.----....(((.......)))...)))))))))........(((((((.....)))))))))))))).)). ( -26.70, z-score = -0.68, R) >droYak2.chr2R 8760914 109 - 21139217 AUUUUGCUGCCAUGCAAUUAAUUCGCCUAUCGCCGU-------ACGGGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA .......((.(((((((..((((((.((((((..((-------(((...----.)(....).......))))...))))))...))))))(((((((.....)))))))))))))).)). ( -29.20, z-score = -1.24, R) >droSec1.super_9 151069 101 + 3197100 AUUUUGCUGCCAUGCAAUUAGUUUGCCUA---------------CGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA .......((.(((((((......((((..---------------((((.----....(((.......)))...))))..)))).......(((((((.....)))))))))))))).)). ( -24.30, z-score = -0.65, R) >droSim1.chr2R 15473912 101 + 19596830 AUUUUGCUGCCAUGCAAUUAGUUUGCCUA---------------CGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA .......((.(((((((......((((..---------------((((.----....(((.......)))...))))..)))).......(((((((.....)))))))))))))).)). ( -24.30, z-score = -0.65, R) >droWil1.scaffold_181141 5129443 84 + 5303230 -UUUCUUUUCCAGGCAAUUAAAUUGCUUAU--------------CGCAA-----CGAGCUC--ACAAAGUGCA--------------AUUUUCAAUCAAAUUGAUUGAAUUGCAUGACAA -..........(((((((...)))))))..--------------.((..-----...))..--.....(((((--------------((..((((((.....)))))))))))))..... ( -18.10, z-score = -1.68, R) >droVir3.scaffold_12823 628098 84 - 2474545 AUUUUUGUUCCAUGCAAUUAACUC-CUUAU--------------CGCAA-----CGAGC---GGCAAA-UGCAUUCG------------CUUCAAUCAAUUUGAUUGAAUUGCAUGACCC ..........(((((((((.....-.....--------------.....-----.((((---(((...-.))...))------------)))(((((.....)))))))))))))).... ( -20.50, z-score = -2.39, R) >droGri2.scaffold_15245 16205767 85 + 18325388 AUAUAUUUUCCAUGCAAUUAAAUC-CUUAU--------------CGCAA-----CGAGC----GUCGAAUGCAUUUU-----------CAUUCACUCAAUUUGAUUGAAUUGCAUGACAC ..........(((((((((.((((-.....--------------(((..-----...))----)..(((((......-----------))))).........)))).))))))))).... ( -20.00, z-score = -3.05, R) >consensus AUUUUGCUUCCAUGCAAUUAAUUCGCCUAUCG_C__________CGAGA____ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAA ..........(((((((........................................((...........))..................(((((((.....)))))))))))))).... (-14.25 = -14.14 + -0.11)


| Location | 16,819,861 – 16,819,970 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Shannon entropy | 0.48076 |
| G+C content | 0.38826 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -13.45 |
| Energy contribution | -13.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.961213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16819861 109 - 21146708 UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UUUCGU-------ACGGCGAUAGGCGAACUAAUUGCAUGGCAGCAAAAU ((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))...... ( -34.40, z-score = -2.81, R) >droPer1.super_4 4947497 108 + 7162766 UUGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUACCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCG--------UUGCCGAUAAGCGAGUUAAUUGCAUAGAAACAAAAA ...(((((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((--------....)))..))))))))..)))))).))........ ( -26.40, z-score = -1.94, R) >dp4.chr3 11446372 108 + 19779522 UUGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUGCCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCG--------UUGCCGAUAAGCGAGUUAAUUGCAUAGAAACAAAAA ...(((((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((--------....)))..))))))))..)))))).))........ ( -26.40, z-score = -1.51, R) >droAna3.scaffold_13266 16118502 120 + 19884421 UUGUCAUGCAAUUCAAUCAAUUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCCGGUUGGGUCUCGUUGGGCGAUAGGCGAUAGGCGAGUUAAUUGCAUGGAAGCAAAAA ((((((((((((((((((.....)))))))(((((...(((((((..(((.(((....((((((.....)).))))..))).).))..)))).))))))))..)))))))...))))... ( -37.00, z-score = -1.68, R) >droEre2.scaffold_4845 10961928 109 - 22589142 UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCGU-------ACGGCGUUAGGCGAACUAAUUGCAUGGCAGCAAAAU ((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))...... ( -34.40, z-score = -2.81, R) >droYak2.chr2R 8760914 109 + 21139217 UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCCCGU-------ACGGCGAUAGGCGAAUUAAUUGCAUGGCAGCAAAAU ((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))...... ( -34.40, z-score = -2.72, R) >droSec1.super_9 151069 101 - 3197100 UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCG---------------UAGGCAAACUAAUUGCAUGGCAGCAAAAU ((((((((((((((((((.....))))))).......((((((.(((((..(((.....)))..)))----))..)---------------).))))......)))))))))))...... ( -30.90, z-score = -3.13, R) >droSim1.chr2R 15473912 101 - 19596830 UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCG---------------UAGGCAAACUAAUUGCAUGGCAGCAAAAU ((((((((((((((((((.....))))))).......((((((.(((((..(((.....)))..)))----))..)---------------).))))......)))))))))))...... ( -30.90, z-score = -3.13, R) >droWil1.scaffold_181141 5129443 84 - 5303230 UUGUCAUGCAAUUCAAUCAAUUUGAUUGAAAAU--------------UGCACUUUGU--GAGCUCG-----UUGCG--------------AUAAGCAAUUUAAUUGCCUGGAAAAGAAA- ...(((.(((((((((((.....))))))((((--------------(((...(((.--.(.....-----)..))--------------)...))))))).))))).)))........- ( -20.40, z-score = -1.57, R) >droVir3.scaffold_12823 628098 84 + 2474545 GGGUCAUGCAAUUCAAUCAAAUUGAUUGAAG------------CGAAUGCA-UUUGCC---GCUCG-----UUGCG--------------AUAAG-GAGUUAAUUGCAUGGAACAAAAAU (..(((((((((((((((.....)))))))(------------((((....-)))))(---((...-----..)))--------------.....-.......))))))))..)...... ( -24.30, z-score = -2.10, R) >droGri2.scaffold_15245 16205767 85 - 18325388 GUGUCAUGCAAUUCAAUCAAAUUGAGUGAAUG-----------AAAAUGCAUUCGAC----GCUCG-----UUGCG--------------AUAAG-GAUUUAAUUGCAUGGAAAAUAUAU ...((((((((((.((((...(((((((.((.-----------...)).)))))))(----((...-----..)))--------------.....-)))).))))))))))......... ( -26.10, z-score = -3.40, R) >consensus UUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU____UCUCG__________G_CGAUAGGCGAAUUAAUUGCAUGGAAGCAAAAU ...(((((((((((((((.....))))))).............................................................(((.....))).))))))))......... (-13.45 = -13.34 + -0.12)



| Location | 16,819,862 – 16,819,971 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.48013 |
| G+C content | 0.38934 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -14.25 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16819862 109 + 21146708 UUUUGCUGCCAUGCAAUUAGUUCGCCUAUCGCCGU-------ACGAAA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA ......((.(((((((.......(((.((((..((-------((....----...............))))...)))))))........(((((((.....)))))))))))))).)).. ( -27.21, z-score = -1.05, R) >droPer1.super_4 4947498 108 - 7162766 UUUUGUUUCUAUGCAAUUAACUCGCUUAUCGGCAA--------CGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGUAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACAAA .((((((...((((((((.(((((....(((....--------)))..----.)))))..((((...(((.((........))))).))))(((((.....))))))))))))))))))) ( -27.70, z-score = -2.01, R) >dp4.chr3 11446373 108 - 19779522 UUUUGUUUCUAUGCAAUUAACUCGCUUAUCGGCAA--------CGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGCAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACAAA .((((((...((((((((.(((((....(((....--------)))..----.)))))..((((....(((........))).....))))(((((.....))))))))))))))))))) ( -27.90, z-score = -1.94, R) >droAna3.scaffold_13266 16118503 120 - 19884421 UUUUGCUUCCAUGCAAUUAACUCGCCUAUCGCCUAUCGCCCAACGAGACCCAACCGGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAAUUGAUUGAAUUGCAUGACAAA .........(((((((..((((((.((((((.....(((.....(((.((.....))))).......)))....))))))...))))))(((((((.....))))))))))))))..... ( -31.30, z-score = -2.86, R) >droEre2.scaffold_4845 10961929 109 + 22589142 UUUUGCUGCCAUGCAAUUAGUUCGCCUAACGCCGU-------ACGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA ......((.(((((((..............(((((-------.((((.----....(((.......)))...)))))))))........(((((((.....)))))))))))))).)).. ( -26.70, z-score = -0.75, R) >droYak2.chr2R 8760915 109 - 21139217 UUUUGCUGCCAUGCAAUUAAUUCGCCUAUCGCCGU-------ACGGGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA ......((.(((((((..((((((.((((((..((-------(((...----.)(....).......))))...))))))...))))))(((((((.....)))))))))))))).)).. ( -29.20, z-score = -1.32, R) >droSec1.super_9 151070 101 + 3197100 UUUUGCUGCCAUGCAAUUAGUUUGCCUA---------------CGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA ......((.(((((((......((((..---------------((((.----....(((.......)))...))))..)))).......(((((((.....)))))))))))))).)).. ( -24.30, z-score = -0.72, R) >droSim1.chr2R 15473913 101 + 19596830 UUUUGCUGCCAUGCAAUUAGUUUGCCUA---------------CGAGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA ......((.(((((((......((((..---------------((((.----....(((.......)))...))))..)))).......(((((((.....)))))))))))))).)).. ( -24.30, z-score = -0.72, R) >droWil1.scaffold_181141 5129444 84 + 5303230 -UUCUUUUCCAGGCAAUUAAAUUGCUUAU--------------CGCAA-----CGAGCUCA--CAAAGUGCA--------------AUUUUCAAUCAAAUUGAUUGAAUUGCAUGACAAA -.........(((((((...)))))))..--------------.((..-----...))...--....(((((--------------((..((((((.....)))))))))))))...... ( -18.10, z-score = -1.70, R) >droVir3.scaffold_12823 628099 84 - 2474545 UUUUUGUUCCAUGCAAUUAACUC-CUUAU--------------CGCAA-----CGAGC---GGCAAA-UGCAUUCG------------CUUCAAUCAAUUUGAUUGAAUUGCAUGACCCG .........(((((((((.....-.....--------------.....-----.((((---(((...-.))...))------------)))(((((.....))))))))))))))..... ( -20.50, z-score = -2.37, R) >droGri2.scaffold_15245 16205768 85 + 18325388 UAUAUUUUCCAUGCAAUUAAAUC-CUUAU--------------CGCAA-----CGAGC----GUCGAAUGCAUUUU-----------CAUUCACUCAAUUUGAUUGAAUUGCAUGACACA .........(((((((((.((((-.....--------------(((..-----...))----)..(((((......-----------))))).........)))).)))))))))..... ( -20.00, z-score = -3.07, R) >consensus UUUUGCUUCCAUGCAAUUAAUUCGCCUAUCG_C__________CGAGA____ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACAAA .........(((((((........................................((...........))..................(((((((.....))))))))))))))..... (-14.25 = -14.14 + -0.11)

| Location | 16,819,862 – 16,819,971 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.50 |
| Shannon entropy | 0.48013 |
| G+C content | 0.38934 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -13.45 |
| Energy contribution | -13.34 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16819862 109 - 21146708 UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UUUCGU-------ACGGCGAUAGGCGAACUAAUUGCAUGGCAGCAAAA .((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))..... ( -34.60, z-score = -2.83, R) >droPer1.super_4 4947498 108 + 7162766 UUUGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUACCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCG--------UUGCCGAUAAGCGAGUUAAUUGCAUAGAAACAAAA (((((.(((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((--------....)))..))))))))..))))))....))))). ( -26.90, z-score = -2.04, R) >dp4.chr3 11446373 108 + 19779522 UUUGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUGCCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCG--------UUGCCGAUAAGCGAGUUAAUUGCAUAGAAACAAAA (((((.(((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((--------....)))..))))))))..))))))....))))). ( -26.90, z-score = -1.61, R) >droAna3.scaffold_13266 16118503 120 + 19884421 UUUGUCAUGCAAUUCAAUCAAUUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCCGGUUGGGUCUCGUUGGGCGAUAGGCGAUAGGCGAGUUAAUUGCAUGGAAGCAAAA (((((((((((((((((((.....)))))))(((((...(((((((..(((.(((....((((((.....)).))))..))).).))..)))).))))))))..)))))))...))))). ( -37.90, z-score = -1.91, R) >droEre2.scaffold_4845 10961929 109 - 22589142 UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCGU-------ACGGCGUUAGGCGAACUAAUUGCAUGGCAGCAAAA .((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))..... ( -34.60, z-score = -2.82, R) >droYak2.chr2R 8760915 109 + 21139217 UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCCCGU-------ACGGCGAUAGGCGAAUUAAUUGCAUGGCAGCAAAA .((((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......-------)))))....))))......)))))))))))..... ( -34.60, z-score = -2.75, R) >droSec1.super_9 151070 101 - 3197100 UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCG---------------UAGGCAAACUAAUUGCAUGGCAGCAAAA .((((((((((((((((((.....))))))).......((((((.(((((..(((.....)))..)))----))..)---------------).))))......)))))))))))..... ( -31.10, z-score = -3.15, R) >droSim1.chr2R 15473913 101 - 19596830 UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCUCG---------------UAGGCAAACUAAUUGCAUGGCAGCAAAA .((((((((((((((((((.....))))))).......((((((.(((((..(((.....)))..)))----))..)---------------).))))......)))))))))))..... ( -31.10, z-score = -3.15, R) >droWil1.scaffold_181141 5129444 84 - 5303230 UUUGUCAUGCAAUUCAAUCAAUUUGAUUGAAAAU--------------UGCACUUUG--UGAGCUCG-----UUGCG--------------AUAAGCAAUUUAAUUGCCUGGAAAAGAA- ....(((.(((((((((((.....))))))((((--------------(((...(((--..(.....-----)..))--------------)...))))))).))))).))).......- ( -20.40, z-score = -1.50, R) >droVir3.scaffold_12823 628099 84 + 2474545 CGGGUCAUGCAAUUCAAUCAAAUUGAUUGAAG------------CGAAUGCA-UUUGCC---GCUCG-----UUGCG--------------AUAAG-GAGUUAAUUGCAUGGAACAAAAA .(..(((((((((((((((.....)))))))(------------((((....-)))))(---((...-----..)))--------------.....-.......))))))))..)..... ( -24.60, z-score = -2.08, R) >droGri2.scaffold_15245 16205768 85 - 18325388 UGUGUCAUGCAAUUCAAUCAAAUUGAGUGAAUG-----------AAAAUGCAUUCGAC----GCUCG-----UUGCG--------------AUAAG-GAUUUAAUUGCAUGGAAAAUAUA (((.((((((((((.((((...(((((((.((.-----------...)).)))))))(----((...-----..)))--------------.....-)))).))))))))))...))).. ( -26.20, z-score = -3.33, R) >consensus UUUGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU____UCUCG__________G_CGAUAGGCGAAUUAAUUGCAUGGAAGCAAAA ....(((((((((((((((.....))))))).............................................................(((.....))).))))))))........ (-13.45 = -13.34 + -0.12)
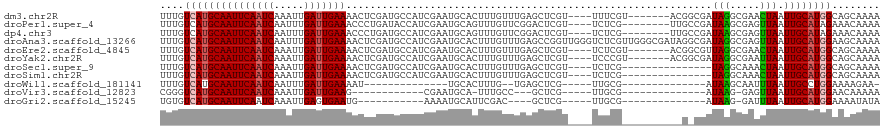

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:36 2011