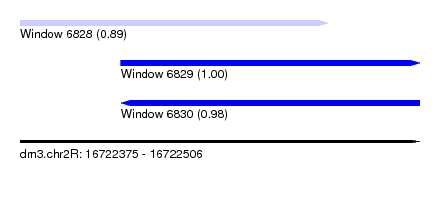
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,722,375 – 16,722,506 |
| Length | 131 |
| Max. P | 0.996848 |

| Location | 16,722,375 – 16,722,476 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Shannon entropy | 0.31982 |
| G+C content | 0.40643 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -18.38 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.894451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16722375 101 + 21146708 -------GAUAUAUUCUGAGGUUUCCAGAUGAGUCGAGAUAACAA-----GUAUUCGAGGUGGCGAAAGCCAACUUCUUCAAAUGCAUAAGCUGAUUAAAAUGUAAGAGACCA -------............((((((((....((((.((.......-----(((((.((((((((....))))....)))).))))).....))))))....))...)))))). ( -25.10, z-score = -1.95, R) >droSim1.chr2R 15375846 101 + 19596830 -------GAUAUAUUCUGAGGUUUCCAGCUGAGUCGAGAUAACAA-----GUAGUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAUAUGAUUAACAUGUAUAUGACCA -------............((((....((....(((((((.....-----...)))((((((((....))).))))).))))..))...(((((.....)))))....)))). ( -27.80, z-score = -1.86, R) >droSec1.super_9 48044 96 + 3197100 -------GAUAUAUUCUGAGGUUUCCAGCUGAGUCGAGAUAACAA-----GUAUUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAU-----UAACAUGUAUAUGACCA -------............((((....((((.((.......))..-----((((((((((((((....))).))))...))))))).....-----...)).))....)))). ( -24.70, z-score = -1.40, R) >droYak2.chr2R 8660802 113 - 21139217 CAUAUUCUGAAUACUCUCAGCUUUCUGGCUGAGUCGAGAUUACUAUACAUGUAUUCGGGGUGGCGAAAGCCAAUUUCUGCGAAUGCAUAAGCUGAUUAAAGGGUAUAUGACAU .(((((((.......(((((((....)))))))....(((((((....(((((((((.((((((....))))....)).))))))))).)).)))))..)))))))....... ( -36.30, z-score = -3.42, R) >consensus _______GAUAUAUUCUGAGGUUUCCAGCUGAGUCGAGAUAACAA_____GUAUUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAGCUGAUUAAAAUGUAUAUGACCA ................................((((......).......((((((((((((((....))))....))))))))))......................))).. (-18.38 = -19.00 + 0.62)

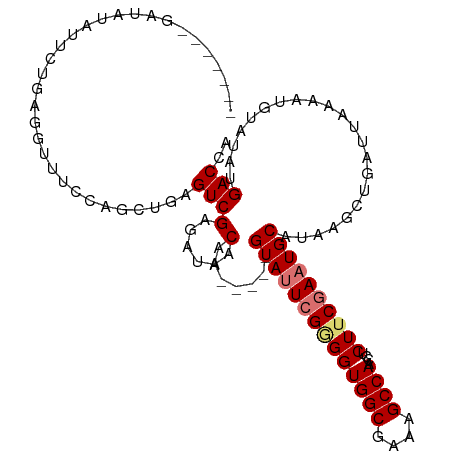
| Location | 16,722,408 – 16,722,506 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.03 |
| Shannon entropy | 0.50741 |
| G+C content | 0.37091 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -16.16 |
| Energy contribution | -17.28 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.996848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16722408 98 + 21146708 -----AACAAGUAUUCGAGGUGGCGAAAGCCAACUUCUUCAAAUGCAUAAGCUGAUUAAAAUGUAAGAGAC----------CAACCAAAACAUUCAAAGAAACUUAAAAAUCU---- -----...((((.(((((((((((....))).)))))......(((((............)))))......----------.................)))))))........---- ( -17.90, z-score = -2.08, R) >droSim1.chr2R 15375879 98 + 19596830 -----AACAAGUAGUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAUAUGAUUAACAUGUAUAUGAC----------CAACAAAAACACUCAAAGAAACUUAAAUGUUU---- -----...((((..((((((((((....))).))))).(((.((((((..((....))..)))))).))).----------.................)).))))........---- ( -21.60, z-score = -2.34, R) >droSec1.super_9 48077 93 + 3197100 -----AACAAGUAUUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUA-----AUUAACAUGUAUAUGAC----------CAACAAAAACACUCAAAGAAACUUAAAUGUUU---- -----...((((.(((((((((((....))).))))).(((.((((((.-----......)))))).))).----------.................)))))))........---- ( -23.10, z-score = -3.69, R) >droYak2.chr2R 8660842 103 - 21139217 UACUAUACAUGUAUUCGGGGUGGCGAAAGCCAAUUUCUGCGAAUGCAUAAGCUGAUUAAAGGGUAUAUGAC----------AUAGCAAAACAUUCACAGAAACUUAAAUGGUU---- ...........((((..((.((((....)))).((((((.(((((((((.(((........))).))))..----------.........))))).))))))))..))))...---- ( -24.90, z-score = -2.30, R) >droEre2.scaffold_4845 10862294 103 + 22589142 --------------UCGGGGUGGCGAAAGCCGACUUCUUCGAAUGCAUAAACUGAUUAAGGUGUUUACAGCAGUAUUCUAUCAAGCUGGAUAUAUUCGGCGGCAUGAAUGAUCGUAA --------------..((((((((....))).)))))..(((.(((.(((((..........)))))..))).(((((......((((((....)))))).....))))).)))... ( -26.90, z-score = -0.75, R) >consensus _____AACAAGUAUUCGGGGUGGCGAAAGCCAACUUCUUCGAAUGCAUAAGCUGAUUAAAAUGUAUAUGAC__________CAACCAAAACAUUCAAAGAAACUUAAAUGUUU____ ..........((((((((((((((....))))....))))))))))....................................................................... (-16.16 = -17.28 + 1.12)

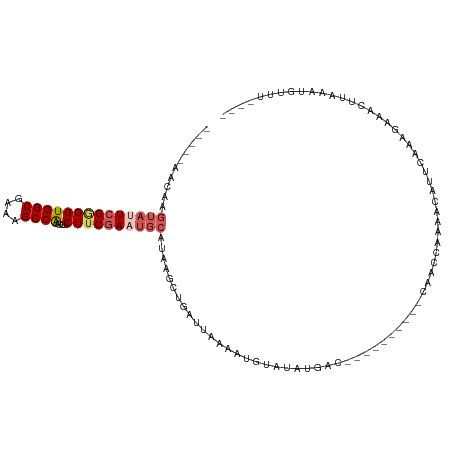
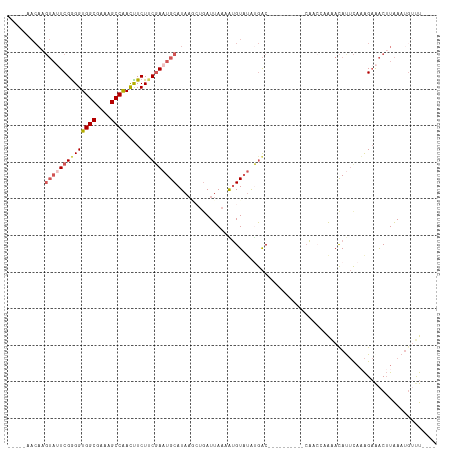
| Location | 16,722,408 – 16,722,506 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 69.03 |
| Shannon entropy | 0.50741 |
| G+C content | 0.37091 |
| Mean single sequence MFE | -22.76 |
| Consensus MFE | -11.81 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
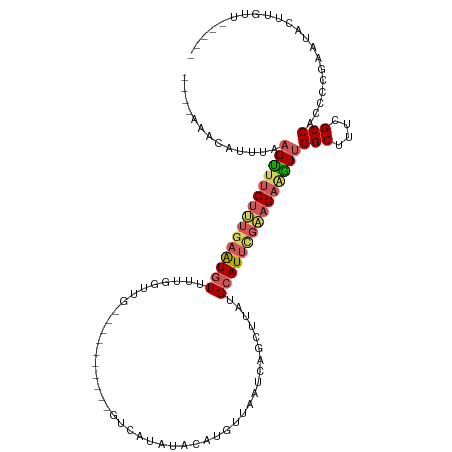
>dm3.chr2R 16722408 98 - 21146708 ----AGAUUUUUAAGUUUCUUUGAAUGUUUUGGUUG----------GUCUCUUACAUUUUAAUCAGCUUAUGCAUUUGAAGAAGUUGGCUUUCGCCACCUCGAAUACUUGUU----- ----.......((((((((((..(((((...(((((----------((.............)))))))...)))))..)))))(.((((....)))).)......)))))..----- ( -24.52, z-score = -2.75, R) >droSim1.chr2R 15375879 98 - 19596830 ----AAACAUUUAAGUUUCUUUGAGUGUUUUUGUUG----------GUCAUAUACAUGUUAAUCAUAUUAUGCAUUCGAAGAGGUUGGCUUUCGCCACCCCGACUACUUGUU----- ----.((((....(((((((((((((((......((----------(((((....)))...))))......)))))))))))((.((((....)))).)).))))...))))----- ( -24.40, z-score = -2.65, R) >droSec1.super_9 48077 93 - 3197100 ----AAACAUUUAAGUUUCUUUGAGUGUUUUUGUUG----------GUCAUAUACAUGUUAAU-----UAUGCAUUCGAAGAGGUUGGCUUUCGCCACCCCGAAUACUUGUU----- ----.......(((((.(((((((((((....((((----------(.(((....))))))))-----...)))))))))))((.((((....)))).)).....)))))..----- ( -24.80, z-score = -3.08, R) >droYak2.chr2R 8660842 103 + 21139217 ----AACCAUUUAAGUUUCUGUGAAUGUUUUGCUAU----------GUCAUAUACCCUUUAAUCAGCUUAUGCAUUCGCAGAAAUUGGCUUUCGCCACCCCGAAUACAUGUAUAGUA ----.........(((((((((((((((...(((((----------...............)).)))....)))))))))))))))(((....)))..................... ( -24.56, z-score = -3.28, R) >droEre2.scaffold_4845 10862294 103 - 22589142 UUACGAUCAUUCAUGCCGCCGAAUAUAUCCAGCUUGAUAGAAUACUGCUGUAAACACCUUAAUCAGUUUAUGCAUUCGAAGAAGUCGGCUUUCGCCACCCCGA-------------- .................(((((...((((......))))(((...(((.((((((..........)))))))))))).......)))))..............-------------- ( -15.50, z-score = 0.61, R) >consensus ____AAACAUUUAAGUUUCUUUGAAUGUUUUGGUUG__________GUCAUAUACAUGUUAAUCAGCUUAUGCAUUCGAAGAAGUUGGCUUUCGCCACCCCGAAUACUUGUU_____ .............(((((((((((((((...........................................)))))))))))))))(((....)))..................... (-11.81 = -12.53 + 0.72)

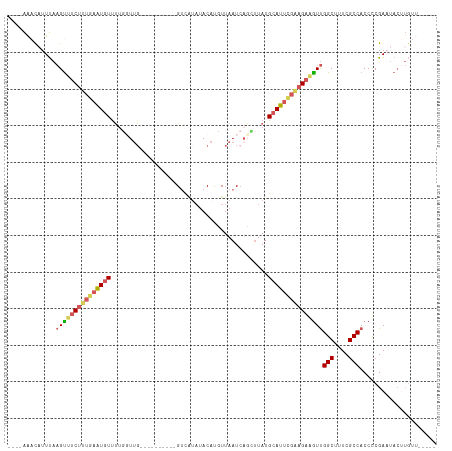
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:27 2011