
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,699,415 – 16,699,511 |
| Length | 96 |
| Max. P | 0.615891 |

| Location | 16,699,415 – 16,699,511 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 59.49 |
| Shannon entropy | 0.78832 |
| G+C content | 0.41176 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -6.86 |
| Energy contribution | -5.97 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.615891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
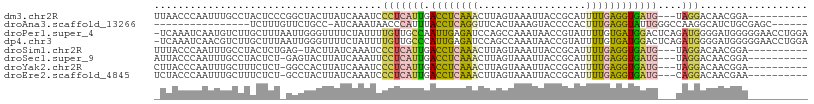
>dm3.chr2R 16699415 96 + 21146708 UUAACCCAAUUUGCCUACUCCCGGCUACUUAUCAAAUCCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG---UAGGACAACGGA---------- .....((.....(((.......)))...........(((..((((.((((((((....(((...))).....))))))))))))---..)))....)).---------- ( -21.00, z-score = -2.00, R) >droAna3.scaffold_13266 15998268 86 - 19884421 ----------------UCUUUGUUCUGCC-AUCAAAUAACCCAUUUACCUCAGGUUCACUAAAGUACCCCACUUUGAGGUAUUGGGCCAAGGCAUCUGCGAGC------ ----------------..(((((..((((-.........((((..((((((((((.(......).)))......))))))).))))....))))...))))).------ ( -21.42, z-score = -1.26, R) >droPer1.super_4 4821163 108 - 7162766 -UCAAAUCAAUGUCUUGCUUUAAUUGGGUUUUCUAUUUUGUUGCCAAUUGAGAUCCAGCCAAAUAACCGUAUUUUGUGAUGGACUCAGAUGGGGAUGGGGGAACCUGGA -......................(..((((..((.....(((.(((.(((((.((((.(((((.........)))).).))))))))).))).))).))..))))..). ( -29.60, z-score = -0.60, R) >dp4.chr3 11321183 108 - 19779522 -UCAAAUCAACGUCUUGCUUUAAUUGGGUUUUCUAUUUUGUUGCCCAUUGAGAUCCAGCCAAAUAACCGUAUUUUGUGAUGGACUCAGAUGGGGAUGGGGGAACCUGGA -......................(..((((..((.......(.(((((((((.((((.(((((.........)))).).)))))))).))))).)..))..))))..). ( -29.30, z-score = -0.26, R) >droSim1.chr2R 15351268 95 + 19596830 UUUACCCAAUUUGCCUACUCUGAG-UACUUAUCAAAUCCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG---UAGGACAACGGA---------- .....((...((((((((.(((((-(.....((((........))))......))))))...((((((.(.....).)))))))---)))).))).)).---------- ( -20.10, z-score = -1.78, R) >droSec1.super_9 25494 95 + 3197100 AUUACCCAAUUUGCCUACUCU-GAGUACUUAUCAAAUUCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG---UAGGACAACGGA---------- .....((...((((((((.((-((((.....((((........))))......))))))...((((((.(.....).)))))))---)))).))).)).---------- ( -20.10, z-score = -1.79, R) >droYak2.chr2R 8637076 95 - 21139217 CUUACCCAAUUUGCUUUCUCU-GGCCACUUAUCAAAUCCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG---UAGGACAACGGA---------- .....((.....(((......-)))...........(((..((((.((((((((....(((...))).....))))))))))))---..)))....)).---------- ( -19.30, z-score = -1.20, R) >droEre2.scaffold_4845 10839399 95 + 22589142 UCUACCCAAUUUGCUUUCUCU-GCCUACUUAUCAAAUCCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG---CAGGACAACGAA---------- ............((.......-))..............(((((((.((((((((....(((...))).....))))))))))))---.)))........---------- ( -15.00, z-score = -1.01, R) >consensus _UUACCCAAUUUGCUUACUCU_GGCUACUUAUCAAAUCCCUCAUUGACCUCAAACUUAGUAAAUUACCGCAUUUUGAGGUGAUG___UAGGACAACGGA__________ ......................................(((((((.((((((((..................))))))))))))....))).................. ( -6.86 = -5.97 + -0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:20 2011