
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,683,679 – 16,683,796 |
| Length | 117 |
| Max. P | 0.567153 |

| Location | 16,683,679 – 16,683,796 |
|---|---|
| Length | 117 |
| Sequences | 7 |
| Columns | 122 |
| Reading direction | forward |
| Mean pairwise identity | 70.00 |
| Shannon entropy | 0.59241 |
| G+C content | 0.40098 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -6.92 |
| Energy contribution | -8.23 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.59 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.26 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
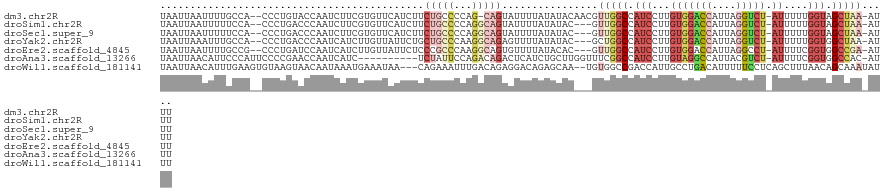
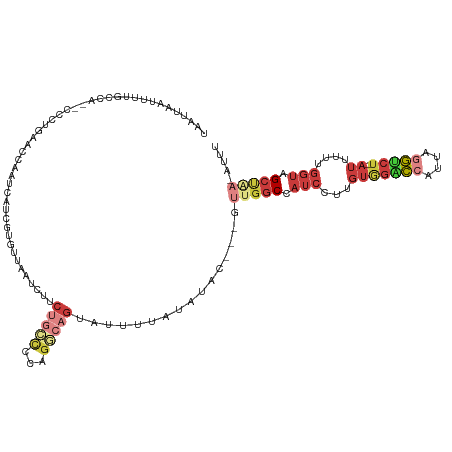
>dm3.chr2R 16683679 117 + 21146708 UAAUUAAUUUUGCCA--CCCUGUACCAAUCUUCGUGUUCAUCUUCUGCCCCAG-CAGUAUUUUAUAUACAACGUUGGCCAUCCUUGUGGACCAUUAGGUCU-AUUUUUGGUAGCUAA-AUUU .....(((((.((.(--((.....(((((.((.((((.......((((....)-)))........)))))).)))))........(((((((....)))))-))....))).)).))-))). ( -25.56, z-score = -2.24, R) >droSim1.chr2R 15334660 115 + 19596830 UAAUUAAUUUUUCCA--CCCUGACCCAAUCUUCGUGUUCAUCUUCUGCCCCAGGCAGUAUUUUAUAUAC---GUUGGCCAUCCUUGUGGACCAUUAGGUCU-AUUUUUGGUAGCUAA-AUUU ...............--...............(((((.......(((((...)))))........))))---)(((((...((..(((((((....)))))-))....))..)))))-.... ( -25.76, z-score = -2.15, R) >droSec1.super_9 9957 115 + 3197100 UAAUUAAUUUUUCCA--CCCUGACCCAAUCUUCGUGUUCAUCUUCUGCCCCAGGCAGUAUUUUAUAUAC---GUUGGCCAUCCUUGUGGACCAUUAGGUCU-AUUUUUGGUAGCUAA-AUUU ...............--...............(((((.......(((((...)))))........))))---)(((((...((..(((((((....)))))-))....))..)))))-.... ( -25.76, z-score = -2.15, R) >droYak2.chr2R 8620983 115 - 21139217 UAAUUAAAUUUGCCA--CCCUGACCCAAUCAUCUUGUUAUUCUGCUGCCCAAGGCAGAGUUUUAUAUAC---GCUGGCCAUCCUUGUGGACCAUUAGGUCU-AUUUUUGGUGGCUAA-AUUU .....((((((((((--((.....(((.......((..((((((((......))))))))..)).....---..)))........(((((((....)))))-))....)))))).))-)))) ( -33.54, z-score = -2.75, R) >droEre2.scaffold_4845 10823534 115 + 22589142 UAAUUAAUUUUGCCG--CCCUGAUCCAAUCAUCUUGUUAUUCUCCCGCCCAAGGCAGUGUUUUAUACAC---GUUGGCCAUCCUUGUGGACCAUUAGGCCU-AUUUUCGGUGGCCGA-AUUU .....(((((.((((--((.((((...))))....................((((.((((.....))))---..(((((((....)))).)))....))))-......)))))).))-))). ( -29.40, z-score = -1.46, R) >droAna3.scaffold_13266 15982140 110 - 19884421 UAAUUAACAUUCCCAUUCCCCGAACCAAUCAUC----------UCUAUUCCAGACAGACUCAUCUGCUUGGUUUCGGCCAUCCUUGUAGGCCAUUACGUCU-AUUUUCGGUGGCCAC-AUUU .....................(((((((.....----------(((.....)))((((....)))).))))))).(((((((...((((((......))))-))....)))))))..-.... ( -26.10, z-score = -3.06, R) >droWil1.scaffold_181141 452758 117 - 5303230 UAAUUAACAUUUGAAGUGUAAGUAACAAUAAAUGAAAUAA---CAGAAAUUUGACAGAGGACAGAGCAA--UGUGGCCGACCAUUGCCUGACAUUUUUCCUCAGCUUUAACAGCAAAUAUUU ........(((((..((..((((..(((....((......---)).....)))...((((((((.((((--((.(.....)))))))))).......))))).))))..))..))))).... ( -22.51, z-score = -1.65, R) >consensus UAAUUAAUUUUGCCA__CCCUGAACCAAUCAUCGUGUUAAUCUUCUGCCCCAGGCAGUAUUUUAUAUAC___GUUGGCCAUCCUUGUGGACCAUUAGGUCU_AUUUUUGGUAGCUAA_AUUU ............................................(((((...)))))...............((((.(((.......(((((....)))))......)))))))........ ( -6.92 = -8.23 + 1.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:18 2011