| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,681,667 – 16,681,765 |
| Length | 98 |
| Max. P | 0.550656 |

| Location | 16,681,667 – 16,681,765 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Shannon entropy | 0.45084 |
| G+C content | 0.48416 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -14.09 |
| Energy contribution | -13.35 |
| Covariance contribution | -0.74 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.550656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
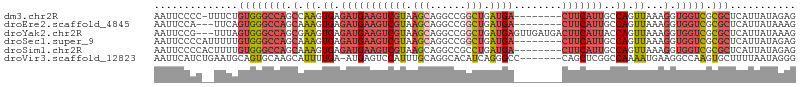
>dm3.chr2R 16681667 98 + 21146708 AAUUCCCC-UUUCUGUGGGCCAGCCAAGUGAGAUGAAGUCGUAAGCAGGCCGGCUGAUGA--------CUUCAUUGCCAGUUAAAGGUGGUCGCGCUCAUUAUAGAG .......(-(((..(((((((.(((......(((((((((((.(((......))).))))--------)))))))(((.......)))))).).))))))...)))) ( -32.50, z-score = -1.68, R) >droEre2.scaffold_4845 10821625 96 + 22589142 AAUUCCA---UUCAGUGGGCCAGCAAAGUGAGAUGAAGUCGUAAGCAGGCCGGCUGAUGA--------CUUCAUUGCCAGUUAAAGGUGGUCGCGCUCAUUAUAAAG .......---...(((((((((.(.((.((.(((((((((((.(((......))).))))--------)))))))..)).))...).))))).)))).......... ( -31.50, z-score = -2.43, R) >droYak2.chr2R 8618948 104 - 21139217 AAUUCCG---UUUAGUGGGCCAGCGAAGUGAGAUGAAGUCGUAAGCAGGCCGGCUGAUGAGUUGAUGACUUCAUUACCAGUUAAAGGUGGUCGCGCUCAUUAUAAAG .......---..((((((((..((((.....(((((((((((.(((......))).........))))))))))).(((.(....).)))))))))))))))..... ( -30.50, z-score = -1.53, R) >droSec1.super_9 7369 99 + 3197100 AAUUCCCCAUUUUUGUGGGCCAGCAAAGUGAGAUGAAGUCGUAAGCAGGCCGGCUGAUGA--------CUUCAUUGCCAGUUAAAGGUGGUCGCGCUCAUUAUAGAG ..............((((((((.(.((.((.(((((((((((.(((......))).))))--------)))))))..)).))...).))))).)))........... ( -30.70, z-score = -1.53, R) >droSim1.chr2R 15332736 99 + 19596830 AAUUCCCCACUUUUGUGGGCCAGCAAAGUGAGAUGAAGUCGUAAGCAGGCCGCCUGAUGA--------CUUCAUUGCCAGUUAAAGGUGGUCGCGCUCAUUAUAGAG .....(((((....))))).......(((((((((....)))..((.((((((((..(((--------((........))))).)))))))))).))))))...... ( -32.80, z-score = -2.26, R) >droVir3.scaffold_12823 436359 99 - 2474545 AAUUCAUCUGAAUGCAGUGCAAGCAUUUUGA-AUGAGUCCAUUUGCAGGCACAUCAGGGCC-------CAGCUCGGCCAAAAUGAAGGCCAAGUGCUUUUAAUAGGG .(((((((.((((((.......)))))).).-))))))((..(((.((((((....((((.-------..))))((((........))))..)))))).)))..)). ( -31.20, z-score = -1.29, R) >consensus AAUUCCCC__UUUUGUGGGCCAGCAAAGUGAGAUGAAGUCGUAAGCAGGCCGGCUGAUGA________CUUCAUUGCCAGUUAAAGGUGGUCGCGCUCAUUAUAGAG ..............((((((..((....((..(((....)))...))(((((.(((((((..........))))).........)).)))))))))))))....... (-14.09 = -13.35 + -0.74)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:15 2011