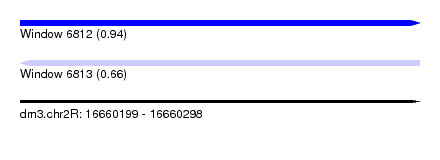
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,660,199 – 16,660,298 |
| Length | 99 |
| Max. P | 0.938137 |

| Location | 16,660,199 – 16,660,298 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.17 |
| Shannon entropy | 0.33056 |
| G+C content | 0.53742 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -19.45 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.938137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
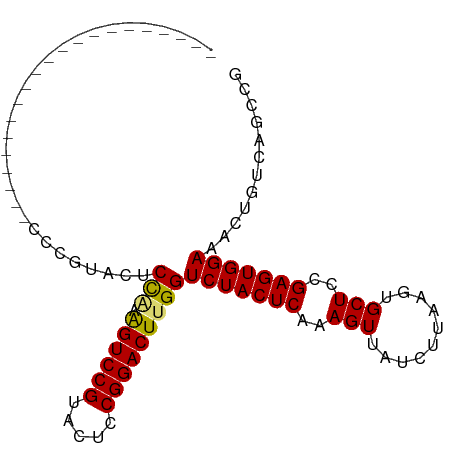
>dm3.chr2R 16660199 99 + 21146708 CCCGUGCUCCAAGGUCCAUCGUCCGGAGUCUGUAGUCCGUACACCGGACUUGGUCUACUCAAAGUUAUCUUAAGUGCUCCGAGUGGAAACUGUCAGCCG ..((.(((...((..(((..((((((.((.((.....)).)).)))))).)))(((((((..(((..........)))..)))))))..))...))))) ( -28.50, z-score = -1.26, R) >droSim1.chr2R 15316036 78 + 19596830 ---------------------CCCGUGCUCCAAAGUCCGUACUCCGGACUUGGUCUACUCAAAGUUAUCUUAAGUGCUCCGAGUGGAAACUGUCAGCCG ---------------------..((.(((((((.(((((.....)))))))))(((((((..(((..........)))..))))))).......))))) ( -23.70, z-score = -2.92, R) >droSec1.super_1 14207451 78 + 14215200 ---------------------CCCGUGCUCCAAAGUCCGUACUCCGGACUUGGUCUACUCAAAGUUAUCUUAAGUGCUCCGAGUGGAAACUGUCAGCCG ---------------------..((.(((((((.(((((.....)))))))))(((((((..(((..........)))..))))))).......))))) ( -23.70, z-score = -2.92, R) >droYak2.chr2R 8602266 85 - 21139217 --------------GCCGUGACCCGAAGUCCUUGGUCCGGAGCUCGGACUUGGUCUACUCAAAGUUAUCUUAAGUGCUCCGAGUGGAAACUGUCAGCCG --------------.(((.((((..........))))))).(((..(((....(((((((..(((..........)))..)))))))....)))))).. ( -24.50, z-score = -0.42, R) >consensus _____________________CCCGUACUCCAAAGUCCGUACUCCGGACUUGGUCUACUCAAAGUUAUCUUAAGUGCUCCGAGUGGAAACUGUCAGCCG .............................(((.((((((.....)))))))))(((((((..(((..........)))..)))))))............ (-19.45 = -19.20 + -0.25)

| Location | 16,660,199 – 16,660,298 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.17 |
| Shannon entropy | 0.33056 |
| G+C content | 0.53742 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.93 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.664297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
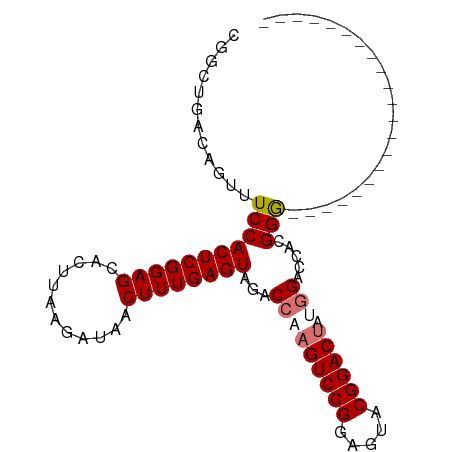
>dm3.chr2R 16660199 99 - 21146708 CGGCUGACAGUUUCCACUCGGAGCACUUAAGAUAACUUUGAGUAGACCAAGUCCGGUGUACGGACUACAGACUCCGGACGAUGGACCUUGGAGCACGGG (((((..(((..((((.((((((((((((((.....)))))))......((((((.....))))))...).))))))....))))..))).))).)).. ( -32.50, z-score = -1.53, R) >droSim1.chr2R 15316036 78 - 19596830 CGGCUGACAGUUUCCACUCGGAGCACUUAAGAUAACUUUGAGUAGACCAAGUCCGGAGUACGGACUUUGGAGCACGGG--------------------- (((((.......((.((((((((............)))))))).))(((((((((.....))))).))))))).))..--------------------- ( -25.90, z-score = -2.22, R) >droSec1.super_1 14207451 78 - 14215200 CGGCUGACAGUUUCCACUCGGAGCACUUAAGAUAACUUUGAGUAGACCAAGUCCGGAGUACGGACUUUGGAGCACGGG--------------------- (((((.......((.((((((((............)))))))).))(((((((((.....))))).))))))).))..--------------------- ( -25.90, z-score = -2.22, R) >droYak2.chr2R 8602266 85 + 21139217 CGGCUGACAGUUUCCACUCGGAGCACUUAAGAUAACUUUGAGUAGACCAAGUCCGAGCUCCGGACCAAGGACUUCGGGUCACGGC-------------- ..((((............(((((((((((((.....))))))).(((...)))...))))))((((..........)))).))))-------------- ( -24.10, z-score = -0.19, R) >consensus CGGCUGACAGUUUCCACUCGGAGCACUUAAGAUAACUUUGAGUAGACCAAGUCCGGAGUACGGACUAUGGACCACGGG_____________________ ...(((......((.((((((((............)))))))).))(((((((((.....)))))).)))....)))...................... (-18.92 = -19.93 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:13 2011