| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,653,028 – 16,653,105 |
| Length | 77 |
| Max. P | 0.983040 |

| Location | 16,653,028 – 16,653,105 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.50527 |
| G+C content | 0.51710 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.88 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.983040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
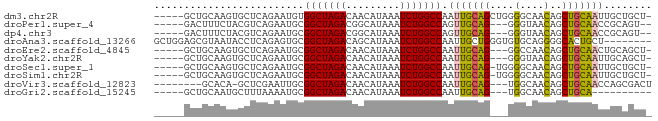
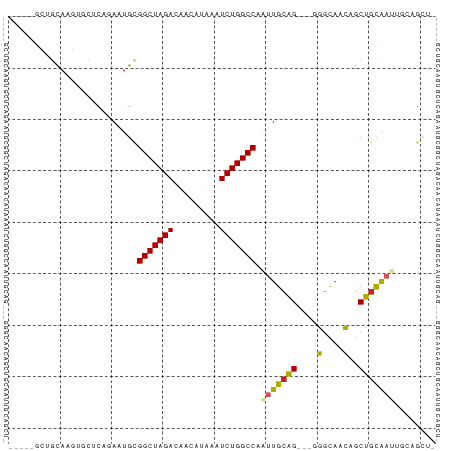
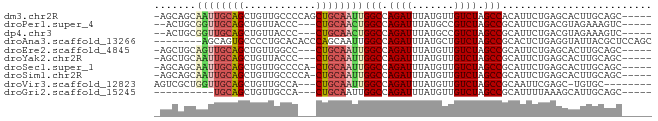
>dm3.chr2R 16653028 77 + 21146708 -----GCUGCAAGUGCUCAGAAUGUGGCUAGACAACAUAAAUCUGGCCAAUUGCAGCUGGGGCAACAGCUGCAAUUGCUGCU- -----((.((((............((((((((.........)))))))).(((((((((......))))))))))))).)).- ( -32.80, z-score = -2.70, R) >droPer1.super_4 4783279 73 - 7162766 -----GACUUUCUACGUCAGAAUGCGGCUAGACGGCAUAAAUCUGGCCAGUUGCAG---GGGUAACAGCUGCAACCGCAGU-- -----(((.......)))....((((((((((.........))))))).(((((((---.(....)..))))))).)))..-- ( -25.10, z-score = -1.85, R) >dp4.chr3 11283395 73 - 19779522 -----GACUUUCUACGUCAGAAUGCGGCUAGACGGCAUAAAUCUGGCCAGUUGCAG---GGGUAACAGCUGCAACCGCAGU-- -----(((.......)))....((((((((((.........))))))).(((((((---.(....)..))))))).)))..-- ( -25.10, z-score = -1.85, R) >droAna3.scaffold_13266 15954161 75 - 19884421 GCUGGAGCGUAAUACCUCAGAGUGCGGCUAGACAGCAUAAAUCUGGCCAAUUGCUGGGUGUGCAGGGGCACUGCU-------- ((....))...(((((.((((((..(((((((.........))))))).))).))))))))((((.....)))).-------- ( -26.10, z-score = -0.29, R) >droEre2.scaffold_4845 10797644 74 + 22589142 -----GCUGCAAGUGCUCAGAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG---GGCCAACAGCUGCAACUGCAGCU- -----.((((((..((.......))(((((((.........)))))))..))))))---.......((((((....))))))- ( -28.40, z-score = -1.92, R) >droYak2.chr2R 8595039 74 - 21139217 -----GCUGCAAGUGCUCAGAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG---GGGUAACAGCUGCAAUUGCAGCU- -----(((((....((.......))(((((((.........)))))))((((((((---.(....)..))))))))))))).- ( -29.20, z-score = -2.60, R) >droSec1.super_1 14200811 76 + 14215200 -----GCUGCAAGUGCUCAGAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG-UGGGGCAACAGCUGCAAUUGCUGCU- -----((.((....((.......))(((((((.........)))))))((((((((-(..(....).))))))))))).)).- ( -29.20, z-score = -1.75, R) >droSim1.chr2R 15308871 76 + 19596830 -----GCUGCAAGUGCUCAGAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG-UGGGGCAACAGCUGCAAUUGCUGCU- -----((.((....((.......))(((((((.........)))))))((((((((-(..(....).))))))))))).)).- ( -29.20, z-score = -1.75, R) >droVir3.scaffold_12823 405466 71 - 2474545 --------GCACA-GCUCGAAUUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG---UGGCAACAGCUGCAACCAGCGACU --------.....-.......(((((((((((.........)))))))..((((((---((....)).))))))...)))).. ( -24.40, z-score = -2.48, R) >droGri2.scaffold_15245 16014207 65 + 18325388 -----GCUGCAAUGCUUUAAAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG---UGGCAACAGCUGCA---------- -----((((((((((........))(((((((.........))))))).)))))))---).(((.....))).---------- ( -23.90, z-score = -2.60, R) >consensus _____GCUGCAAGUGCUCAGAAUGCGGCUAGACAACAUAAAUCUGGCCAAUUGCAG___GGGCAACAGCUGCAAUUGCAGCU_ .........................(((((((.........))))))).(((((((....(....)..)))))))........ (-16.02 = -15.88 + -0.14)
| Location | 16,653,028 – 16,653,105 |
|---|---|
| Length | 77 |
| Sequences | 10 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 75.18 |
| Shannon entropy | 0.50527 |
| G+C content | 0.51710 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.13 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
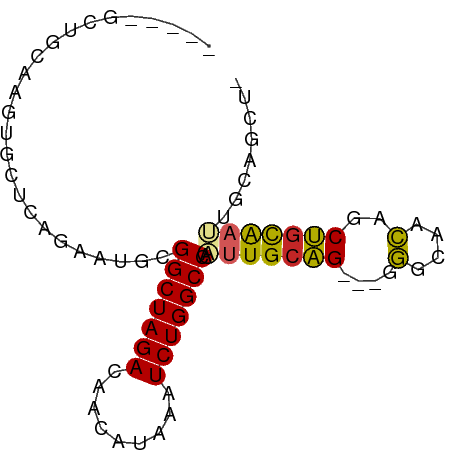
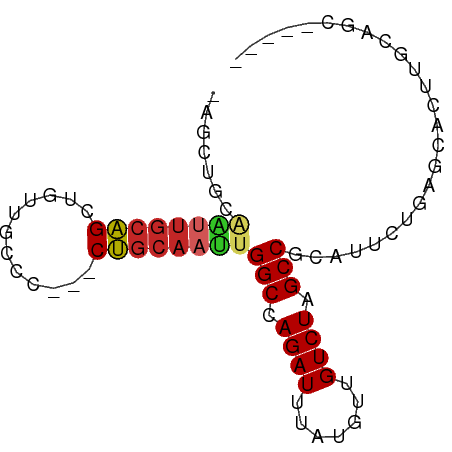
>dm3.chr2R 16653028 77 - 21146708 -AGCAGCAAUUGCAGCUGUUGCCCCAGCUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCACAUUCUGAGCACUUGCAGC----- -.((((((((((((((((......))))))))))))((((((...((((.(.....).)))))))).))..))))...----- ( -30.80, z-score = -2.75, R) >droPer1.super_4 4783279 73 + 7162766 --ACUGCGGUUGCAGCUGUUACCC---CUGCAACUGGCCAGAUUUAUGCCGUCUAGCCGCAUUCUGACGUAGAAAGUC----- --.((((((((((((.........---))))))).(((.((((.......)))).))).........)))))......----- ( -22.50, z-score = -1.59, R) >dp4.chr3 11283395 73 + 19779522 --ACUGCGGUUGCAGCUGUUACCC---CUGCAACUGGCCAGAUUUAUGCCGUCUAGCCGCAUUCUGACGUAGAAAGUC----- --.((((((((((((.........---))))))).(((.((((.......)))).))).........)))))......----- ( -22.50, z-score = -1.59, R) >droAna3.scaffold_13266 15954161 75 + 19884421 --------AGCAGUGCCCCUGCACACCCAGCAAUUGGCCAGAUUUAUGCUGUCUAGCCGCACUCUGAGGUAUUACGCUCCAGC --------(((.((((....))))((((((....((((.((((.......)))).))))....))).))).....)))..... ( -21.80, z-score = -0.75, R) >droEre2.scaffold_4845 10797644 74 - 22589142 -AGCUGCAGUUGCAGCUGUUGGCC---CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUCUGAGCACUUGCAGC----- -((((((....)))))).......---((((((..(((.((((.......)))).)))((.......))..)))))).----- ( -25.50, z-score = -0.38, R) >droYak2.chr2R 8595039 74 + 21139217 -AGCUGCAAUUGCAGCUGUUACCC---CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUCUGAGCACUUGCAGC----- -((((((....)))))).......---((((((..(((.((((.......)))).)))((.......))..)))))).----- ( -25.50, z-score = -1.91, R) >droSec1.super_1 14200811 76 - 14215200 -AGCAGCAAUUGCAGCUGUUGCCCCA-CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUCUGAGCACUUGCAGC----- -.(((((((((((((.((......))-)))))))))((((((...((((.(.....).)))))))).))..))))...----- ( -24.00, z-score = -0.66, R) >droSim1.chr2R 15308871 76 - 19596830 -AGCAGCAAUUGCAGCUGUUGCCCCA-CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUCUGAGCACUUGCAGC----- -.(((((((((((((.((......))-)))))))))((((((...((((.(.....).)))))))).))..))))...----- ( -24.00, z-score = -0.66, R) >droVir3.scaffold_12823 405466 71 + 2474545 AGUCGCUGGUUGCAGCUGUUGCCA---CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAAUUCGAGC-UGUGC-------- ....((..(((((((.((....))---)))))))..))(((.(((..(((((......)))))..))))-))...-------- ( -21.30, z-score = -0.01, R) >droGri2.scaffold_15245 16014207 65 - 18325388 ----------UGCAGCUGUUGCCA---CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUUUAAAGCAUUGCAGC----- ----------....((....))..---(((((((.(((.((((.......)))).)))((........))))))))).----- ( -18.10, z-score = -0.26, R) >consensus _AGCUGCAAUUGCAGCUGUUGCCC___CUGCAAUUGGCCAGAUUUAUGUUGUCUAGCCGCAUUCUGAGCACUUGCAGC_____ .......((((((((............))))))))(((.((((.......)))).)))......................... (-14.00 = -14.13 + 0.13)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:10 2011