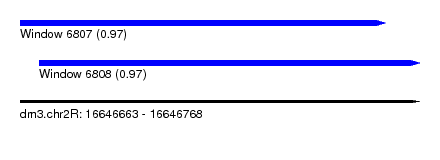
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,646,663 – 16,646,768 |
| Length | 105 |
| Max. P | 0.974084 |

| Location | 16,646,663 – 16,646,759 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 70.73 |
| Shannon entropy | 0.53201 |
| G+C content | 0.40053 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -11.18 |
| Energy contribution | -10.53 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.93 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
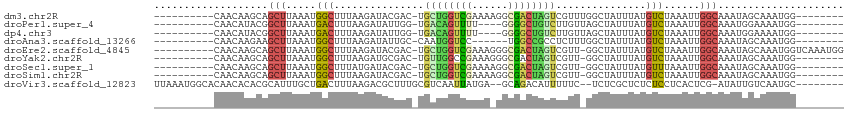
>dm3.chr2R 16646663 96 + 21146708 ----------CAACAAGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUUUGGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGG-------- ----------...(((((((((.....))))............-.((((((((......)))))))).)))))((((((..((((......))))))))))......-------- ( -27.80, z-score = -2.80, R) >droPer1.super_4 4776924 92 - 7162766 ----------CAACAUACGGCUUAAAUGACUUUAAGAUAUUGG-UGACAGUUUU----GGGGCUGUCUUGUUAGCUAUUUAUGUCUAAAUUGGCAAAUGGAAAAUGG-------- ----------.((((.(((((((............((.((((.-...)))).))----.)))))))..))))..(((((..((((......))))))))).......-------- ( -18.62, z-score = -1.11, R) >dp4.chr3 11277053 92 - 19779522 ----------CAACAUACGGCUUAAAUGACUUUAAGAUAUUGG-UGACAGUUUU----GGGGCUGUCUUGUUAGCUAUUUAUGUCUAAAUUGGCAAAUGGAAAAUGG-------- ----------.((((.(((((((............((.((((.-...)))).))----.)))))))..))))..(((((..((((......))))))))).......-------- ( -18.62, z-score = -1.11, R) >droAna3.scaffold_13266 15948243 90 - 19884421 ----------CAACAAGAAGCUUAAAUGGCUUUAAGAUAUUGC-CAAUGGUCC------UGGCCGCCUCUUUGGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGG-------- ----------......((((((.....))))))......((((-(((((((..------..)))(((.....))).............))))))))...........-------- ( -24.00, z-score = -1.32, R) >droEre2.scaffold_4845 10791418 103 + 22589142 ----------CAACAAGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAGGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGUCAAAUGG ----------........((((.....))))....(((.((((-.((((((((......)))))))).)))-)((((((..((((......)))))))))).....)))...... ( -30.00, z-score = -2.86, R) >droYak2.chr2R 8588529 95 - 21139217 ----------CAACAAGCAGCUUAAAUGGCUUUAAGAUGCGAC-UGUUGGCCGAAAGGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGG-------- ----------((((..(((.((((((....)))))).)))(((-(((((.((....)).)))).)))))))-)((((((..((((......))))))))))......-------- ( -37.30, z-score = -5.22, R) >droSec1.super_1 14194550 95 + 14215200 ----------CAACAAGCAGCUUAAAUGGCUUUAUGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUU-GGCUAUUUAUGUUUAAAUUGGCAAAUAGCAAAUGG-------- ----------...((.(.((((.....)))).).))...((((-.((((((((......)))))))).)))-)((((((..((((......))))))))))......-------- ( -26.50, z-score = -2.45, R) >droSim1.chr2R 15302481 95 + 19596830 ----------CAACAAGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGG-------- ----------........((((.....))))........((((-.((((((((......)))))))).)))-)((((((..((((......))))))))))......-------- ( -28.30, z-score = -3.04, R) >droVir3.scaffold_12823 395853 102 - 2474545 UUAAAUGGCACAACACACGCAUUUGCUGACUUUAAGACGCUUUGCGUCAAUUAUGA--GCAGACAUUUUUC--UCUCGCUCUCUCCUCACUCG-AUAUUGUCAAUGC-------- .......(((........((....))((((..((.(((((...)))))......((--(((((........--))).))))............-.))..)))).)))-------- ( -19.70, z-score = -0.91, R) >consensus __________CAACAAGCAGCUUAAAUGGCUUUAAGAUACGAC_UGCUGGUCGAAA_GGCGACUAGUCGUU_GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGG________ ...................(((.....(((...............((((((((......))))))))...............)))......)))..................... (-11.18 = -10.53 + -0.65)

| Location | 16,646,668 – 16,646,768 |
|---|---|
| Length | 100 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Shannon entropy | 0.60543 |
| G+C content | 0.42231 |
| Mean single sequence MFE | -28.87 |
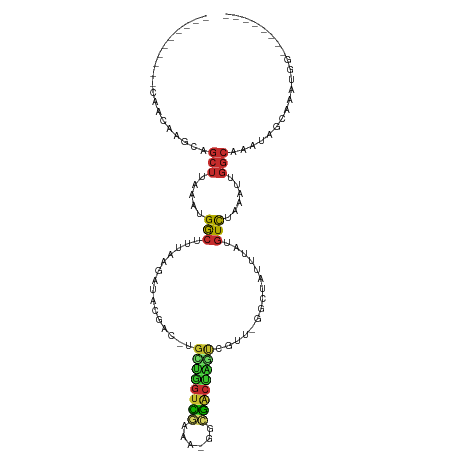
| Consensus MFE | -11.35 |
| Energy contribution | -10.80 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.88 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.974084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
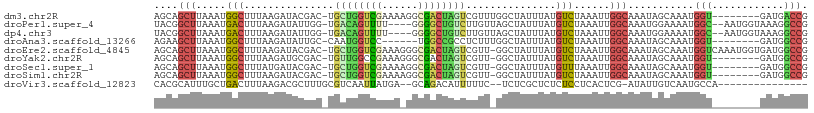
>dm3.chr2R 16646668 100 + 21146708 AGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUUUGGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGU--------GAUGACCG ....((((((((((((.......(((((-((.(.(((.....))).).)))))))..)))))))))((((......))))...)))....(((--------....))). ( -28.60, z-score = -2.05, R) >droPer1.super_4 4776929 102 - 7162766 UACGGCUUAAAUGACUUUAAGAUAUUGG-UGACAGUUUU----GGGGCUGUCUUGUUAGCUAUUUAUGUCUAAAUUGGCAAAUGGAAAAUGGC--AAUGGUAAAGGCCG ..((((((......(((((((((..((.-...)))))))----)))).(..(((((((.(((((..((((......)))))))))....))))--)).)..).)))))) ( -25.60, z-score = -1.80, R) >dp4.chr3 11277058 102 - 19779522 UACGGCUUAAAUGACUUUAAGAUAUUGG-UGACAGUUUU----GGGGCUGUCUUGUUAGCUAUUUAUGUCUAAAUUGGCAAAUGGAAAAUGGC--AAUGGUAAAGGCCG ..((((((......(((((((((..((.-...)))))))----)))).(..(((((((.(((((..((((......)))))))))....))))--)).)..).)))))) ( -25.60, z-score = -1.80, R) >droAna3.scaffold_13266 15948248 94 - 19884421 AGAAGCUUAAAUGGCUUUAAGAUAUUGC-CAAUGGUCC------UGGCCGCCUCUUUGGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGU--------GAUGGCCG .((((((.....))))))........((-((..(((..------..)))(((......((((((..((((......))))))))))....)))--------..)))).. ( -27.00, z-score = -1.10, R) >droEre2.scaffold_4845 10791423 107 + 22589142 AGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAGGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGUCAAAUGGUGAUGGCCG ...((((.....))))........((((-.((((((((......)))))))).)))-)((((((..((((......))))))))))....(((((........))))). ( -34.10, z-score = -2.74, R) >droYak2.chr2R 8588534 99 - 21139217 AGCAGCUUAAAUGGCUUUAAGAUGCGAC-UGUUGGCCGAAAGGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGU--------GAUGGCCG ....((((((((((((.......(((((-(((((.((....)).)))).)))))).-)))))))))((((......))))...)))....(((--------....))). ( -35.70, z-score = -3.34, R) >droSec1.super_1 14194555 99 + 14215200 AGCAGCUUAAAUGGCUUUAUGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUU-GGCUAUUUAUGUUUAAAUUGGCAAAUAGCAAAUGGU--------GAUGGCCG ............((((((((.((.((((-.((((((((......)))))))).)))-)((((((..((((......))))))))))..)).))--------)).)))). ( -31.80, z-score = -2.92, R) >droSim1.chr2R 15302486 99 + 19596830 AGCAGCUUAAAUGGCUUUAAGAUACGAC-UGCUGGUCGAAAAGGCGACUAGUCGUU-GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGU--------GAUGGCCG ............(((((((..((.((((-.((((((((......)))))))).)))-)((((((..((((......))))))))))..))..)--------)).)))). ( -31.30, z-score = -2.60, R) >droVir3.scaffold_12823 395868 89 - 2474545 CACGCAUUUGCUGACUUUAAGACGCUUUGCGUCAAUUAUGA--GCAGACAUUUUUC--UCUCGCUCUCUCCUCACUCG-AUAUUGUCAAUGCCA--------------- ...((((....((((..((.(((((...)))))......((--(((((........--))).))))............-.))..))))))))..--------------- ( -20.10, z-score = -2.02, R) >consensus AGCAGCUUAAAUGGCUUUAAGAUACGAC_UGCUGGUCGAAA_GGCGACUAGUCGUU_GGCUAUUUAUGUCUAAAUUGGCAAAUAGCAAAUGGU________GAUGGCCG ....(((.....(((...............((((((((......))))))))...............)))......)))...........(((............))). (-11.35 = -10.80 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:09 2011