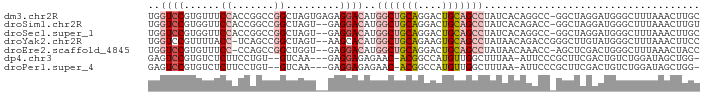
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,633,767 – 16,633,858 |
| Length | 91 |
| Max. P | 0.741169 |

| Location | 16,633,767 – 16,633,858 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 68.24 |
| Shannon entropy | 0.58922 |
| G+C content | 0.55958 |
| Mean single sequence MFE | -32.37 |
| Consensus MFE | -10.02 |
| Energy contribution | -10.23 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
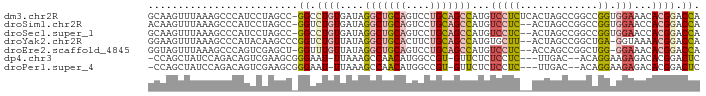
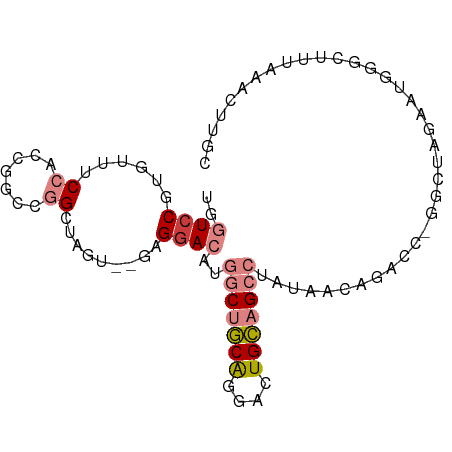
>dm3.chr2R 16633767 91 + 21146708 GCAAGUUUAAAGCCCAUCCUAGCC-GGCCUGUGAUAGGCUGCAGUCCUGCAGCCAUGUCCUCUCACUAGCCGGCCGGUGGAAACACGGACCA ................(((..(((-(((..((((..(((((((....)))))))..(....)))))..))))))..(((....))))))... ( -38.50, z-score = -3.13, R) >droSim1.chr2R 15289356 89 + 19596830 ACAAGUUUAAAGCCCAUCCUAGCC-GGUCUGUGAUAGGCUGCAGUCCUGCAGCCAUGUCCUC--ACUAGCCGGCCGGUGGAACCACGGACCA ................(((..(((-(((..(((((((((((((....))))))).))...))--))..))))))..(((....))))))... ( -35.20, z-score = -2.56, R) >droSec1.super_1 14181663 89 + 14215200 GCAAGUUUAAAGCCCAUCCUAGCC-GGCCUGUGAUAGGCUGCAGUCCUGCAGCCAUGUCCUC--ACUAGCCGGCCGGUGGAACCACGGACCA ................(((..(((-(((..(((((((((((((....))))))).))...))--))..))))))..(((....))))))... ( -36.50, z-score = -2.50, R) >droYak2.chr2R 8575316 89 - 21139217 GGAAGUUUAAAGCCCAUACAAGCCCGGUCUGUUAUAGGCUGCACUUCUGCAGCCAUGUGCUU--ACUAGCCGGCUGA-GGUAAAACGGACCA ((..((((...(((......((((.(((..(((((((((((((....))))))).))))...--))..)))))))..-))).))))...)). ( -28.80, z-score = -0.94, R) >droEre2.scaffold_4845 10778570 88 + 22589142 GGUAGUUUAAAGCCCAGUCGAGCU-GGUUUGUUAUAGGCUGCAGUCCUGCAGCCAUGUCCUC--ACCAGCCGGCUGG-GGAAACACGGACCA (((.((((....((((((((.(((-(((....(((.(((((((....)))))))))).....--)))))))))))))-).))))....))). ( -44.00, z-score = -4.66, R) >dp4.chr3 11263413 84 - 19779522 -CCAGCUAUCCAGACAGUCGAAGCGGGAAU-UUAAAGCCAACAUGGCCGU-GUUCUCUCCUC---UUGAC--ACAGGAAGAGACACGGACUC -..............((((...........-.....(((.....)))(((-((.(((((((.---.....--..))).))))))))))))). ( -21.80, z-score = -0.51, R) >droPer1.super_4 4763388 84 - 7162766 -CCAGCUAUCCAGACAGUCGAAGCGGGAAU-UUAAAGCCAACAUGGCCGU-GUUCUCUCCUC---UUGAC--ACAGGAAGAGACACGGACUC -..............((((...........-.....(((.....)))(((-((.(((((((.---.....--..))).))))))))))))). ( -21.80, z-score = -0.51, R) >consensus GCAAGUUUAAAGCCCAUCCGAGCC_GGUCUGUUAUAGGCUGCAGUCCUGCAGCCAUGUCCUC__ACUAGCCGGCCGGUGGAAACACGGACCA .........................((.........(((((((....)))))))...................(((.........))).)). (-10.02 = -10.23 + 0.21)
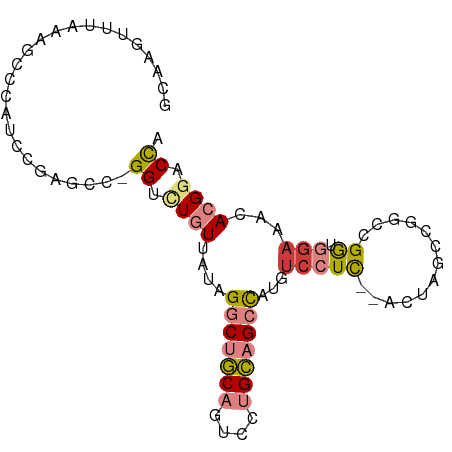
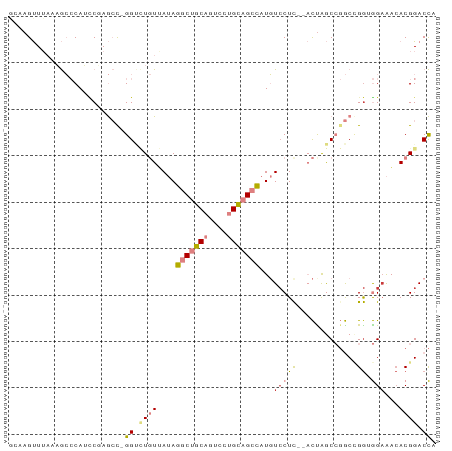
| Location | 16,633,767 – 16,633,858 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 68.24 |
| Shannon entropy | 0.58922 |
| G+C content | 0.55958 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -11.47 |
| Energy contribution | -12.31 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.702250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16633767 91 - 21146708 UGGUCCGUGUUUCCACCGGCCGGCUAGUGAGAGGACAUGGCUGCAGGACUGCAGCCUAUCACAGGCC-GGCUAGGAUGGGCUUUAAACUUGC .((((((....(((...((((((((.((((........(((((((....)))))))..)))).))))-)))).))))))))).......... ( -41.50, z-score = -2.72, R) >droSim1.chr2R 15289356 89 - 19596830 UGGUCCGUGGUUCCACCGGCCGGCUAGU--GAGGACAUGGCUGCAGGACUGCAGCCUAUCACAGACC-GGCUAGGAUGGGCUUUAAACUUGU .(((((((((.....))((((((((.((--((......(((((((....)))))))..)))))).))-))))...))))))).......... ( -39.80, z-score = -2.58, R) >droSec1.super_1 14181663 89 - 14215200 UGGUCCGUGGUUCCACCGGCCGGCUAGU--GAGGACAUGGCUGCAGGACUGCAGCCUAUCACAGGCC-GGCUAGGAUGGGCUUUAAACUUGC .(((((((((.....))((((((((.((--((......(((((((....)))))))..)))).))))-))))...))))))).......... ( -44.20, z-score = -3.40, R) >droYak2.chr2R 8575316 89 + 21139217 UGGUCCGUUUUACC-UCAGCCGGCUAGU--AAGCACAUGGCUGCAGAAGUGCAGCCUAUAACAGACCGGGCUUGUAUGGGCUUUAAACUUCC .(((((((...((.-..((((.(((...--.)))....(((((((....)))))))............)))).))))))))).......... ( -26.80, z-score = -0.18, R) >droEre2.scaffold_4845 10778570 88 - 22589142 UGGUCCGUGUUUCC-CCAGCCGGCUGGU--GAGGACAUGGCUGCAGGACUGCAGCCUAUAACAAACC-AGCUCGACUGGGCUUUAAACUACC .(((....((((.(-((((.((((((((--..(.....(((((((....))))))).....)..)))-))).)).)))))....)))).))) ( -37.90, z-score = -3.20, R) >dp4.chr3 11263413 84 + 19779522 GAGUCCGUGUCUCUUCCUGU--GUCAA---GAGGAGAGAAC-ACGGCCAUGUUGGCUUUAA-AUUCCCGCUUCGACUGUCUGGAUAGCUGG- ..(.((((((((((((((..--....)---).)))))).))-)))).)...(..(((....-..(((.((.......))..))).)))..)- ( -24.60, z-score = -0.55, R) >droPer1.super_4 4763388 84 + 7162766 GAGUCCGUGUCUCUUCCUGU--GUCAA---GAGGAGAGAAC-ACGGCCAUGUUGGCUUUAA-AUUCCCGCUUCGACUGUCUGGAUAGCUGG- ..(.((((((((((((((..--....)---).)))))).))-)))).)...(..(((....-..(((.((.......))..))).)))..)- ( -24.60, z-score = -0.55, R) >consensus UGGUCCGUGUUUCCACCGGCCGGCUAGU__GAGGACAUGGCUGCAGGACUGCAGCCUAUAACAGACC_GGCUAGAAUGGGCUUUAAACUUGC ..((((.........((....)).........))))..(((((((....))))))).................................... (-11.47 = -12.31 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:07 2011