
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,453,412 – 3,453,484 |
| Length | 72 |
| Max. P | 0.899882 |

| Location | 3,453,412 – 3,453,484 |
|---|---|
| Length | 72 |
| Sequences | 13 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 69.92 |
| Shannon entropy | 0.58312 |
| G+C content | 0.52480 |
| Mean single sequence MFE | -16.19 |
| Consensus MFE | -10.90 |
| Energy contribution | -10.50 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.80 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.899882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3453412 72 - 23011544 UCAUCCUGCUCCAUAAUGGAUAAGGCUAGUGCUCGUGCUAGGGCCUCGUCUUCGCUCUGCAAGU-UUUCAAUA-------- ......(((........(((..(((((...((....))...)))))..))).......)))...-........-------- ( -17.56, z-score = -0.61, R) >droSim1.chr2L 3407818 73 - 22036055 UCAUCCUGCUCCAUAAUGGAUAAGGCUAGGGCUCGUGCGAGGGCCUCGUCUUCGCUCUGCAAGUAUUUCAAUC-------- ....((((((((.....)))....)).)))((....((((((((...))))))))...)).............-------- ( -19.30, z-score = -0.38, R) >droSec1.super_129 15489 73 - 51580 UCAUCCUGCUCCAUAAUGGAUAAGGCUAGGGCUCGUGCGAGGGCCUCGUCUUCGCUCUGUAAGUAUUUCAAUC-------- ...(((((((((.....)))....)).))))...(.((((((((...)))))))).)................-------- ( -17.70, z-score = -0.06, R) >droYak2.chr2L 3441244 73 - 22324452 UCAUCCUGCUCCAUGAUUGAUAAGGCUAGGGCUCGUGCCAGCGCCUCGUCUUCGCUCUGCAAGUAUUUAAAUC-------- ......(((....(((..((..((((...(((....)))...))))..)).)))....)))............-------- ( -18.90, z-score = -0.88, R) >droEre2.scaffold_4929 3488631 73 - 26641161 UCAUCCUGCUCCAUGAUUGAUAAGGCCAGCGCUCGUGCGAGGGCCUCGUCUUCGCUCUGUAAGUAUUUAAAAC-------- ......(((....(((..((..(((((..(((....)))..)))))..)).)))....)))............-------- ( -20.90, z-score = -1.53, R) >droAna3.scaffold_12943 2531794 59 + 5039921 UCAUCCUGUUCCAUAAUCGAUAAAGCAAGAGCUCGUGCCAGUGCCUCGUCUUCACUCUG---------------------- ..................((..(.(((...((....))...))).)..)).........---------------------- ( -6.60, z-score = 0.36, R) >dp4.chr4_group3 926328 59 - 11692001 UCAUCCUGUUCCAUUAUUGAUAAGGCUAGCGCCCGUGCCAGAGCCUCGUCUUCAGUCUG---------------------- .((..(((..........((..(((((.(((....)))...)))))..))..)))..))---------------------- ( -13.30, z-score = -1.46, R) >droPer1.super_1 2404781 59 - 10282868 UCAUCCUGUUCCAUUAUUGAUAAGGCUAGCGCCCGUGCCAGAGCCUCGUCUUCAGUCUG---------------------- .((..(((..........((..(((((.(((....)))...)))))..))..)))..))---------------------- ( -13.30, z-score = -1.46, R) >droWil1.scaffold_180772 572838 59 + 8906247 UCAUCCUGUUCCAUAAUGGACAGGGCUAAAGCACGGGCCAGUGCCUCAUCUUCGCUCUG---------------------- ...((((((((......)))))))).....((((......))))...............---------------------- ( -15.20, z-score = -0.72, R) >droGri2.scaffold_15252 13370177 59 - 17193109 UCAUCCUCCUCCAUGAUGGACAGGGCCAAAGCAUGCGCCAAAGCCUCGUCCUCAGUCUG---------------------- .............(((.((((..(((....((....))....)))..))))))).....---------------------- ( -16.10, z-score = -1.45, R) >droMoj3.scaffold_6500 5385773 59 - 32352404 UCAUCUUGUUCCAUAAUUGAUAAAGCCAGAGCAUGUGCCAGGGCUUCGUCCUCAGUCUA---------------------- ..................((..(((((...((....))...)))))..)).........---------------------- ( -11.70, z-score = -0.55, R) >droVir3.scaffold_12963 15941794 59 - 20206255 UCAUCCUGCUCCAUGAUGGACAAAGCUAGCGCAUGCGCCAAAGCCUCGUCCUCAGUCUG---------------------- .............(((.((((...(((.(((....)))...)))...))))))).....---------------------- ( -14.00, z-score = -0.84, R) >anoGam1.chr2R 42446871 81 + 62725911 UCCUCCUCCUCCUGCAUGGACAGGGCGAUCGCACGUGCUAGCGCUUCGUCCUCCGACAUACCGCCCUGGACGAUUGCGAUA ..(.(((..(((.....)))..))).)((((((.(((....))).((((((..((......))....)))))).)))))). ( -25.90, z-score = -1.09, R) >consensus UCAUCCUGCUCCAUAAUGGAUAAGGCUAGCGCUCGUGCCAGGGCCUCGUCUUCACUCUG______________________ ..................(((.(((((...((....))...))))).)))............................... (-10.90 = -10.50 + -0.40)

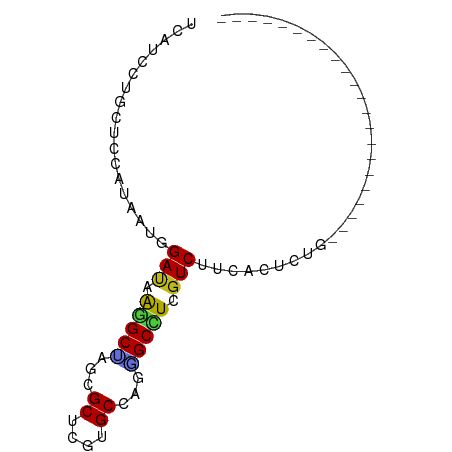
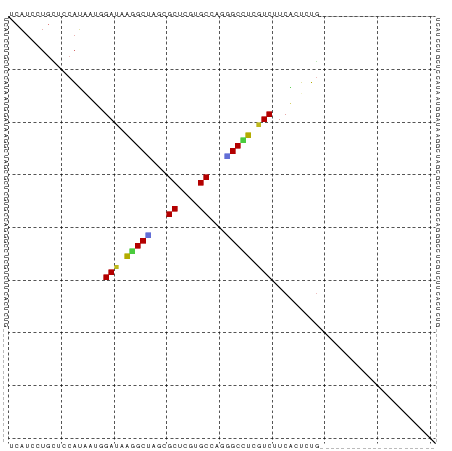
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:55 2011