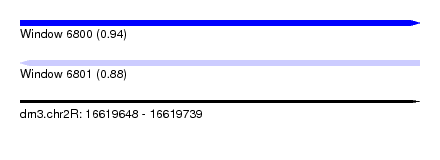
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,619,648 – 16,619,739 |
| Length | 91 |
| Max. P | 0.939354 |

| Location | 16,619,648 – 16,619,739 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.40323 |
| G+C content | 0.41345 |
| Mean single sequence MFE | -21.92 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.21 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.939354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
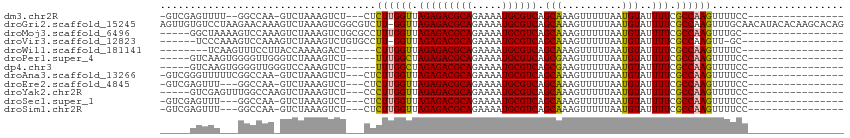
>dm3.chr2R 16619648 91 + 21146708 -GUCGAGUUUU--GGCCAA-GUCUAAAGUCU---CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -...(((.(((--(((...-.......((((---((.......))))))...((((((((((.((........)).))))))))))))))))..))).---------------- ( -23.30, z-score = -2.50, R) >droGri2.scaffold_15245 15978999 113 + 18325388 AGUUGUGUCCUAAGAACAAAGUCUAAAGUCGGCGUCUU-GGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUGCAACAUACACAAGCACAG .((((((..((.(((......)))......((((((((-.....)))))...((((((((((.((........)).))))))))))))).))..)))))).............. ( -26.60, z-score = -1.16, R) >droMoj3.scaffold_6496 25424994 92 - 26866924 -----GGCUAAAAGUCCAAAGUCUAAAGUCUGCGCCUUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUGC----------------- -----(((.((((..((((((.(..........).)))))).....((((((.....))))))..................)))).)))........----------------- ( -20.40, z-score = -0.53, R) >droVir3.scaffold_12823 364553 89 - 2474545 ------UCCCAAAGUCCAAAGUCUAAAGUCUGUGCCUU-GGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUU-GC----------------- ------.........................((..(((-(((.(((((((((.....)))))).(((..........)))..))).))))))..-))----------------- ( -17.90, z-score = -1.26, R) >droWil1.scaffold_181141 4769086 84 + 5303230 --------UCAAGUUUCCUUACCAAAAGACU-----CUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUC----------------- --------....(((((...(((((......-----.)))))..)))))((.((((((((((.((........)).)))))))))))).........----------------- ( -18.30, z-score = -2.03, R) >droPer1.super_4 4753466 88 - 7162766 -----GUCAAGUGGGGUUGGGUCUAAAGUCU-----UUUGGCUAGAGACGCAGAAAAUGCGUCAGCGAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -----.........(((((.((((..((((.-----...))))..)))).))((((((((((.((........)).))))))))))))).........---------------- ( -22.50, z-score = -1.39, R) >dp4.chr3 11253483 88 - 19779522 -----GUCAAGUGGGGUUGGGUCCAAAGUCU-----UUUGGCUAGAGACGCAGAAAAUGCGUCAGCGAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -----((((((.((..(((....)))..)).-----))))))....((((((.....)))))).((((((............))))))..........---------------- ( -23.70, z-score = -1.68, R) >droAna3.scaffold_13266 15924589 93 - 19884421 -GUCGGGUUUUUCGGCCAA-GUCUAAAGUCU---CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -...((((((((.((((((-(..........---..)))))))))))))...((((((((((.((........)).)))))))))).)).........---------------- ( -21.20, z-score = -1.45, R) >droEre2.scaffold_4845 10769171 90 + 22589142 -GUCGAGUUU---GGCCAA-GUCUAAAGUCU---CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -...((..((---(((...-.......((((---((.......))))))...((((((((((.((........)).)))))))))))))))..))...---------------- ( -22.60, z-score = -2.30, R) >droYak2.chr2R 8565701 90 - 21139217 -----GUCGAGUUUGGCCAAGUCUAAAGUCU---CCCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -----(.((((((((((((((..........---..))))))))))((((((.....))))))....................)))).).........---------------- ( -21.40, z-score = -2.13, R) >droSec1.super_1 14172464 90 + 14215200 -GUCGAGUUU---GGCCAA-GUCUAAAGUCU---CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -...((..((---(((...-.......((((---((.......))))))...((((((((((.((........)).)))))))))))))))..))...---------------- ( -22.60, z-score = -2.30, R) >droSim1.chr2R 15280060 90 + 19596830 -GUCGAGUUU---GGCCAA-GUCUAAAGUCU---CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC---------------- -...((..((---(((...-.......((((---((.......))))))...((((((((((.((........)).)))))))))))))))..))...---------------- ( -22.60, z-score = -2.30, R) >consensus _____GGUUU___GGCCAA_GUCUAAAGUCU___CUCUUGGUUAGAGACGCAGAAAAUGCGUCAGCAAAGUUUUUAAUGUAUUUUCGCCAAGUUUUCC________________ ....................................((((((.(((((((((.....)))))).(((..........)))..))).))))))...................... (-15.56 = -15.21 + -0.35)
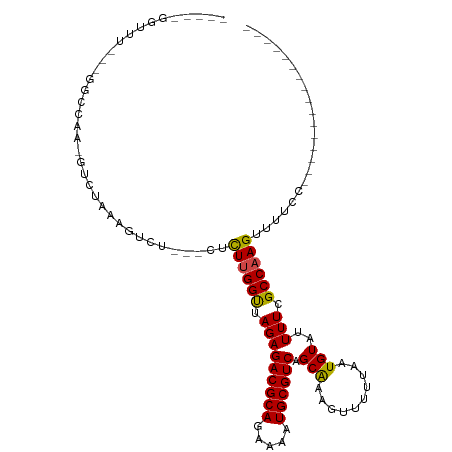
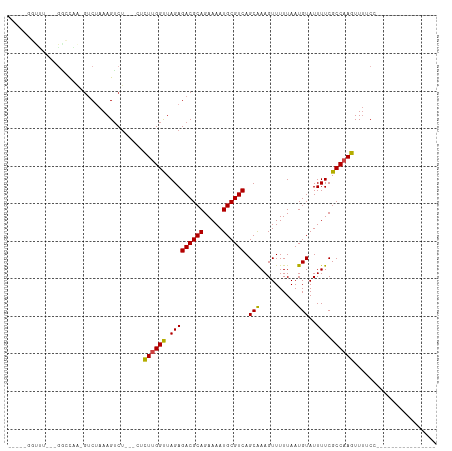
| Location | 16,619,648 – 16,619,739 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Shannon entropy | 0.40323 |
| G+C content | 0.41345 |
| Mean single sequence MFE | -19.19 |
| Consensus MFE | -12.03 |
| Energy contribution | -11.89 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16619648 91 - 21146708 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG---AGACUUUAGAC-UUGGCC--AAAACUCGAC- ----------------.......(((((.((................(((....)))...((((.((((((.......))---))))..)))).-)).)))--))........- ( -18.00, z-score = -1.54, R) >droGri2.scaffold_15245 15978999 113 - 18325388 CUGUGCUUGUGUAUGUUGCAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACC-AAGACGCCGACUUUAGACUUUGUUCUUAGGACACAACU .((((((((.....(((..(((.((((((......................((((((.....))))))(((....-.))))))))).))).)))........)))).))))... ( -24.72, z-score = -1.12, R) >droMoj3.scaffold_6496 25424994 92 + 26866924 -----------------GCAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAAGGCGCAGACUUUAGACUUUGGACUUUUAGCC----- -----------------........(((.((((..((..........(((....)))...(((((((((........)))))))))..........))...)))).)))----- ( -19.10, z-score = -1.04, R) >droVir3.scaffold_12823 364553 89 + 2474545 -----------------GC-AACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACC-AAGGCACAGACUUUAGACUUUGGACUUUGGGA------ -----------------..-.....((((((..............))))))((((((.....)))))).....((-((((..((((........))))..))))))..------ ( -21.74, z-score = -2.61, R) >droWil1.scaffold_181141 4769086 84 - 5303230 -----------------GAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAG-----AGUCUUUUGGUAAGGAAACUUGA-------- -----------------........((((((..............))))))((((((.....))))))....((((((-----(...)))))))(((....)))..-------- ( -19.44, z-score = -2.22, R) >droPer1.super_4 4753466 88 + 7162766 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUCGCUGACGCAUUUUCUGCGUCUCUAGCCAAA-----AGACUUUAGACCCAACCCCACUUGAC----- ----------------((.......((((((..............))))))((((((.....))))))..........-----...........)).............----- ( -15.44, z-score = -1.70, R) >dp4.chr3 11253483 88 + 19779522 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUCGCUGACGCAUUUUCUGCGUCUCUAGCCAAA-----AGACUUUGGACCCAACCCCACUUGAC----- ----------------((.......((((((..............))))))((((((.....)))))).....(((((-----....)))))..)).............----- ( -18.44, z-score = -2.38, R) >droAna3.scaffold_13266 15924589 93 + 19884421 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG---AGACUUUAGAC-UUGGCCGAAAAACCCGAC- ----------------((.....(((((.((................(((....)))...((((.((((((.......))---))))..)))).-)).))))).....))...- ( -19.10, z-score = -1.84, R) >droEre2.scaffold_4845 10769171 90 - 22589142 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG---AGACUUUAGAC-UUGGCC---AAACUCGAC- ----------------.......(((((.((................(((....)))...((((.((((((.......))---))))..)))).-)).)))---)).......- ( -18.00, z-score = -1.55, R) >droYak2.chr2R 8565701 90 + 21139217 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGGG---AGACUUUAGACUUGGCCAAACUCGAC----- ----------------.......(((((.((....................((((((.....))))))(((((....((.---...))))))).)).))))).......----- ( -20.30, z-score = -2.17, R) >droSec1.super_1 14172464 90 - 14215200 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG---AGACUUUAGAC-UUGGCC---AAACUCGAC- ----------------.......(((((.((................(((....)))...((((.((((((.......))---))))..)))).-)).)))---)).......- ( -18.00, z-score = -1.55, R) >droSim1.chr2R 15280060 90 - 19596830 ----------------GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG---AGACUUUAGAC-UUGGCC---AAACUCGAC- ----------------.......(((((.((................(((....)))...((((.((((((.......))---))))..)))).-)).)))---)).......- ( -18.00, z-score = -1.55, R) >consensus ________________GGAAAACUUGGCGAAAAUACAUUAAAAACUUUGCUGACGCAUUUUCUGCGUCUCUAACCAAGAG___AGACUUUAGAC_UUGGCC___AAACC_____ .........................((((((..............))))))((((((.....)))))).............................................. (-12.03 = -11.89 + -0.14)

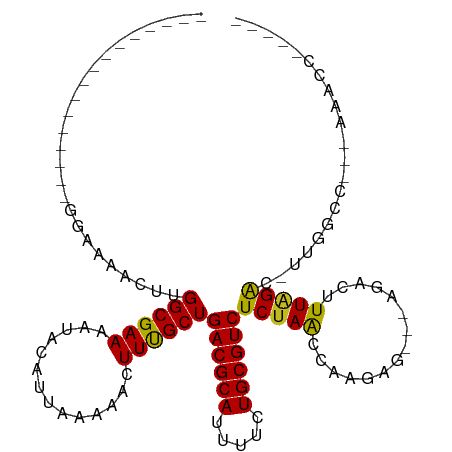
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:03 2011