
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,617,844 – 16,617,943 |
| Length | 99 |
| Max. P | 0.996804 |

| Location | 16,617,844 – 16,617,943 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Shannon entropy | 0.45949 |
| G+C content | 0.48799 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -19.54 |
| Energy contribution | -20.62 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994116 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
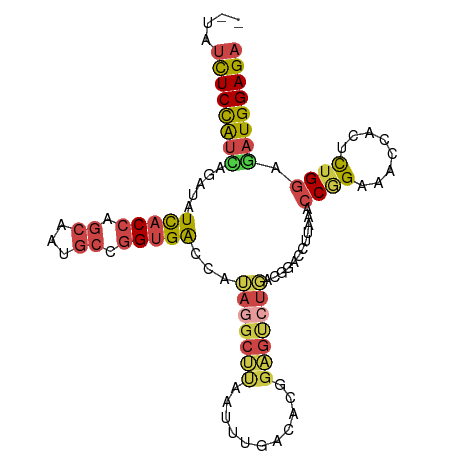
>dm3.chr2R 16617844 99 + 21146708 UCUCCAUCUCCAGAGUGGUUUCCGGUUUAAGGUCCGACAGACUCCGUGUCAAAUUAAGCCUAUGGUCACCGGCAUUGCUGGUGAUAUCUAAUGGAGAUA-- .....(((((((...(((.....(((((((.....((((.......))))...)))))))....(((((((((...)))))))))..))).))))))).-- ( -33.50, z-score = -2.52, R) >droEre2.scaffold_4845 10767371 101 + 22589142 UCUCCAUCUCCAGGGUGGUUUCUGGCUUAAGGUCCGCAAGACUCUUUGUCGGAUUAAGCCUUUGGCCACCGGCAUUGGCGGUGAUAUCUGCUGGAGAUCAU .....((((((..(((((((...((((((..(((((((((....)))).)))))))))))...)))))))......(((((......)))))))))))... ( -42.90, z-score = -3.40, R) >droYak2.chr2R 8563912 89 - 21139217 UCUCCAUCUGCAGGGUGGUUUCGGGUUUUAGGUCCGGA------------GGAUUAAGCUUAUGGCCACCGGCAUUGCUGGUGAUAUCUGAUGGAGAUAAU (((((((((((..(((((((..((((((..(((((...------------)))))))))))..))))))).))).....((......)))))))))).... ( -32.70, z-score = -2.14, R) >droSec1.super_1 14170652 99 + 14215200 UCUCCAUUUCCAGAGUGGUUUCCGGUUUAAGGUCCGUCAGACUCCGUGUCAAAUUAAGCCUAUGGUCACCAGCAUUGCUGGUGAUAUCUAAUGGAGAUA-- ((((((((.(((....(((((.(((........)))...(((.....))).....)))))..)))((((((((...)))))))).....))))))))..-- ( -33.90, z-score = -3.48, R) >droSim1.chr2R 15278231 99 + 19596830 UCUCCAUUUCCAGAGUGGUUUCCGGUUUAAGGUCCGUCAGACUCCGUGUCAAAUUAAGCCUAUGGUCACCAGCAUUGCUGGUGAUAUCUGAUGGAGAUA-- (((((((...((((..(((((.(((........)))...(((.....))).....)))))....(((((((((...))))))))).)))))))))))..-- ( -35.40, z-score = -3.51, R) >droAna3.scaffold_13266 15922903 101 - 19884421 UCUCCAUCUCCAACGUGGUGUCCGGCUUGAGCUCUUCCACGCCCCUAAUGCCAUCGGGCUUAUGAUCUCGGGCAUUUCGGGUAAUAUCUAACAGAACUCAA ......(((.....((((.....(((....)))...))))((((..((((((..((((........))))))))))..))))..........)))...... ( -25.40, z-score = -0.49, R) >consensus UCUCCAUCUCCAGAGUGGUUUCCGGUUUAAGGUCCGCCAGACUCCGUGUCAAAUUAAGCCUAUGGUCACCGGCAUUGCUGGUGAUAUCUAAUGGAGAUA__ ((((((((...............(((((((.........(((.....)))...)))))))....(((((((((...)))))))))....)))))))).... (-19.54 = -20.62 + 1.08)


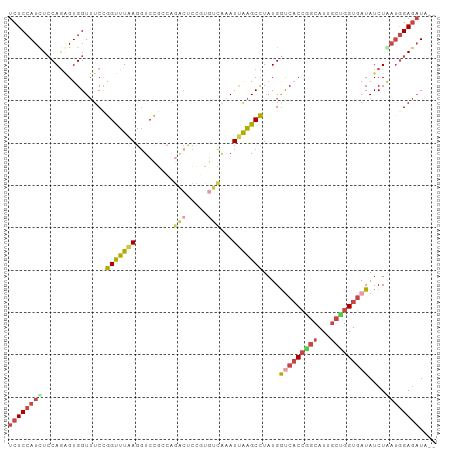
| Location | 16,617,844 – 16,617,943 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Shannon entropy | 0.45949 |
| G+C content | 0.48799 |
| Mean single sequence MFE | -30.89 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.996804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16617844 99 - 21146708 --UAUCUCCAUUAGAUAUCACCAGCAAUGCCGGUGACCAUAGGCUUAAUUUGACACGGAGUCUGUCGGACCUUAAACCGGAAACCACUCUGGAGAUGGAGA --..((((((((.....(((((.((...)).)))))((((((((((...........)))))))).))........(((((......))))).)))))))) ( -32.60, z-score = -2.90, R) >droEre2.scaffold_4845 10767371 101 - 22589142 AUGAUCUCCAGCAGAUAUCACCGCCAAUGCCGGUGGCCAAAGGCUUAAUCCGACAAAGAGUCUUGCGGACCUUAAGCCAGAAACCACCCUGGAGAUGGAGA .(.((((((((.........(((((......))))).....(((((((((((.(((......)))))))..)))))))..........)))))))).)... ( -35.90, z-score = -3.30, R) >droYak2.chr2R 8563912 89 + 21139217 AUUAUCUCCAUCAGAUAUCACCAGCAAUGCCGGUGGCCAUAAGCUUAAUCC------------UCCGGACCUAAAACCCGAAACCACCCUGCAGAUGGAGA ....((((((((.....(((((.((...)).)))))......((.......------------..(((.........)))..........)).)))))))) ( -22.71, z-score = -2.32, R) >droSec1.super_1 14170652 99 - 14215200 --UAUCUCCAUUAGAUAUCACCAGCAAUGCUGGUGACCAUAGGCUUAAUUUGACACGGAGUCUGACGGACCUUAAACCGGAAACCACUCUGGAAAUGGAGA --..((((((((.....((((((((...))))))))((.(((((((...........)))))))..))........(((((......))))).)))))))) ( -34.10, z-score = -4.29, R) >droSim1.chr2R 15278231 99 - 19596830 --UAUCUCCAUCAGAUAUCACCAGCAAUGCUGGUGACCAUAGGCUUAAUUUGACACGGAGUCUGACGGACCUUAAACCGGAAACCACUCUGGAAAUGGAGA --..(((((((......((((((((...))))))))((.(((((((...........)))))))..))........(((((......)))))..))))))) ( -32.60, z-score = -3.55, R) >droAna3.scaffold_13266 15922903 101 + 19884421 UUGAGUUCUGUUAGAUAUUACCCGAAAUGCCCGAGAUCAUAAGCCCGAUGGCAUUAGGGGCGUGGAAGAGCUCAAGCCGGACACCACGUUGGAGAUGGAGA (((((((((......(((..(((..((((((....(((........))))))))).)))..)))..))))))))).(((..(.((.....)).).)))... ( -27.40, z-score = -0.45, R) >consensus __UAUCUCCAUCAGAUAUCACCAGCAAUGCCGGUGACCAUAGGCUUAAUUUGACACGGAGUCUGACGGACCUUAAACCGGAAACCACUCUGGAGAUGGAGA ....((((((((.....(((((.((...)).)))))...(((((((...........)))))))............((((........)))).)))))))) (-18.00 = -18.87 + 0.87)

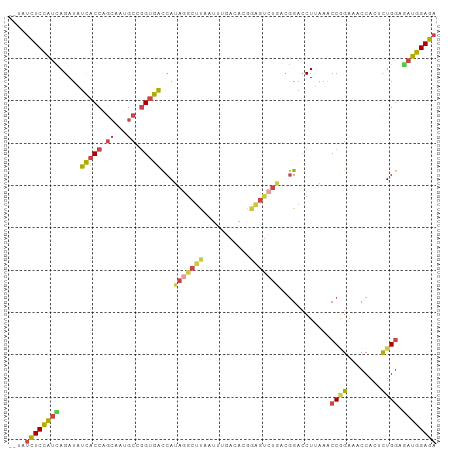
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:39:01 2011