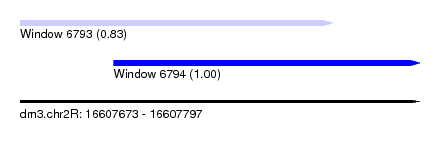
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,607,673 – 16,607,797 |
| Length | 124 |
| Max. P | 0.995525 |

| Location | 16,607,673 – 16,607,770 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.41340 |
| G+C content | 0.48010 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -16.50 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.832250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
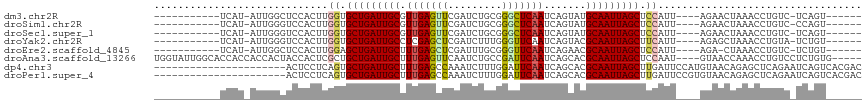
>dm3.chr2R 16607673 97 + 21146708 --------------------UCAUCUAC-UACCACUGGCAGUGUGC-GUUUUCAU-AUUGGCUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAG --------------------........-(((((.((((((((((.-.....)))-))))...)))..)))))((((((((((((((((((.....))))))))......)))))))))) ( -29.60, z-score = -1.51, R) >droSim1.chr2R 15268614 97 + 19596830 --------------------UCAUCUAC-UACCACUACCAGUGUGC-GUUUUCAU-AUUGGGUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAG --------------------........-(((((...((((((((.-.....)))-))))).......)))))((((((((((((((((((.....))))))))......)))))))))) ( -30.30, z-score = -2.27, R) >droSec1.super_1 14160996 97 + 14215200 --------------------UCAUCUAC-UACCACUACCAGUGUGC-GUUGUCAU-AUUGGGUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAG --------------------........-(((((...((((((((.-.....)))-))))).......)))))((((((((((((((((((.....))))))))......)))))))))) ( -30.30, z-score = -1.73, R) >droYak2.chr2R 8553023 99 - 21139217 ------------------UGUCAUCUUC-UACCAAUGGCACUGUGU-GCUUUCAU-AUUGGGUCCACUUGGUGCUGAUUGCCUCGAGCUCGAUCUUUGGGUUCAAUCAGUACGCAAUUAG ------------------..........-..((((((((((...))-))).....-)))))......((((((((((((((((.(((......))).)))..)))))))))).))).... ( -26.80, z-score = -1.21, R) >droEre2.scaffold_4845 10757640 97 + 22589142 --------------------UCAUCUUC-UACCAUUGGCACUGUGU-GGUUUCAU-AUUGGCUCCACUUGGAGCUGAUUGCUUUGAGCUCGAUUUGCGGGUUCAAUCAGAACGCAAUUAG --------------------........-............(((((-(((.....-((..(((((....)))))..)).)))(((((((((.....))))))))).....)))))..... ( -27.40, z-score = -1.37, R) >droAna3.scaffold_13266 15912896 120 - 19884421 GCCCAGCUCCCUCCCACAAGCUGUCUCCCUGGCACUAGCCUGGUAUUGGCACCACCACCACUACCACUCGCUGCUGAUUGCUUUGAGUUCAAUCUGCCGAUUCAAUCAGCACGCAAUUAG ...(((((..........))))).......(((....)))(((((.(((........))).)))))...(((((((((((..(((.((.......)))))..))))))))).))...... ( -35.00, z-score = -3.35, R) >consensus ____________________UCAUCUAC_UACCACUAGCAGUGUGC_GUUUUCAU_AUUGGGUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUACGCAAUUAG .............................(((((..(((...(((.......))).....))).....)))))((((((((.(((((((((.....))))))))).......)))))))) (-16.50 = -16.20 + -0.30)

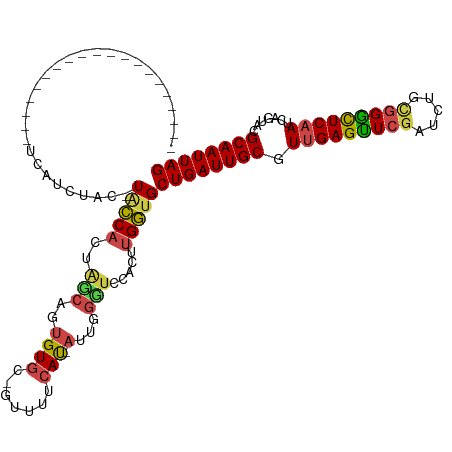
| Location | 16,607,702 – 16,607,797 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 70.21 |
| Shannon entropy | 0.51630 |
| G+C content | 0.45362 |
| Mean single sequence MFE | -29.31 |
| Consensus MFE | -16.07 |
| Energy contribution | -16.01 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16607702 95 + 21146708 -----------UCAU-AUUGGCUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAGCUCCAUU----AGAACUAAACCUGUC-UCAGU------ -----------....-.((((.((....(((.(((((((((((((((((((.....))))))))......))))))))))).)))..----.)).)))).......-.....------ ( -30.80, z-score = -2.72, R) >droSim1.chr2R 15268643 95 + 19596830 -----------UCAU-AUUGGGUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAGCUCCAUU----AGAACUAAACCUGUC-CCAGU------ -----------....-((((((......(((.(((((((((((((((((((.....))))))))......))))))))))).))).(----((........))).)-)))))------ ( -33.70, z-score = -3.46, R) >droSec1.super_1 14161025 95 + 14215200 -----------UCAU-AUUGGGUCCACUUGGUGCUGAUUGCGUUGAGUUCGAUCUGCGGGCUCAAUCAGUAUGCAAUUAGCUCCAUU----AGAACUAAACCUGUC-UCAGU------ -----------....-.((((.((....(((.(((((((((((((((((((.....))))))))......))))))))))).)))..----.)).)))).......-.....------ ( -30.80, z-score = -2.60, R) >droYak2.chr2R 8553054 95 - 21139217 -----------UCAU-AUUGGGUCCACUUGGUGCUGAUUGCCUCGAGCUCGAUCUUUGGGUUCAAUCAGUACGCAAUUAGCUUCAUU----AGAGCUAAACCUGUA-UCUGU------ -----------....-...((((....((((((((((((((((.(((......))).)))..)))))))))).)))(((((((....----.)))))))))))...-.....------ ( -28.10, z-score = -2.08, R) >droEre2.scaffold_4845 10757669 94 + 22589142 -----------UCAU-AUUGGCUCCACUUGGAGCUGAUUGCUUUGAGCUCGAUUUGCGGGUUCAAUCAGAACGCAAUUAGCUCCAUU----AGA-CUAAACCUGUC-UCUGU------ -----------....-............(((((((((((((.(((((((((.....))))))))).......)))))))))))))..----(((-(.......)))-)....------ ( -33.00, z-score = -3.79, R) >droAna3.scaffold_13266 15912936 109 - 19884421 UGGUAUUGGCACCACCACCACUACCACUCGCUGCUGAUUGCUUUGAGUUCAAUCUGCCGAUUCAAUCAGCACGCAAUUAGCUCCAAU----GUAACCAAACCUGUCCUCUGUG----- (((((.(((........))).)))))...(((((((((((..(((.((.......)))))..))))))))).)).............----......................----- ( -26.80, z-score = -2.29, R) >dp4.chr3 11242426 96 - 19779522 ----------------------ACUCCUCAGUGCUGAUUGCUUUGAGCCAAAUCUUUGGAUUCAAUCAGCACGCAAUUAGCUUGAUUCCAUGUAACAGAGCUCAGAAUCAGUCACGAC ----------------------........((((((((...((((((((.......((((.(((...(((.........)))))).)))).......).)))))))))))).)))... ( -25.34, z-score = -1.48, R) >droPer1.super_4 4742431 96 - 7162766 ----------------------ACUCCUCAGUGCUGAUUGCUUUGAGCCAAAUCUUUGGAUUCAAUCAGCACGCAAUUAGCUUGAUUCCGUGUAACAGAGCUCAGAAUCAGUCACGAC ----------------------........((((((((((.......((((....))))...))))))))))((.....))(((((((.(.((......)).).)))))))....... ( -25.90, z-score = -1.43, R) >consensus ___________UCAU_AUUGGGUCCACUUGGUGCUGAUUGCUUUGAGCUCGAUCUGCGGGUUCAAUCAGUACGCAAUUAGCUCCAUU____AGAACUAAACCUGUC_UCAGU______ .............................((.(((((((((.(((((((.........))))))).......))))))))).)).................................. (-16.07 = -16.01 + -0.06)
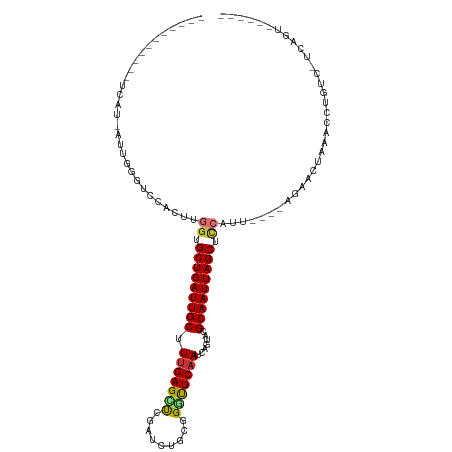
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:58 2011