| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,592,303 – 16,592,400 |
| Length | 97 |
| Max. P | 0.931230 |

| Location | 16,592,303 – 16,592,400 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.13 |
| Shannon entropy | 0.49979 |
| G+C content | 0.50682 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -19.63 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
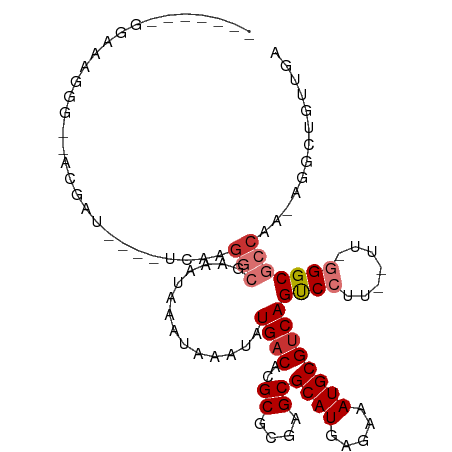
>dm3.chr2R 16592303 97 - 21146708 -------GGAAAGGGACGCGAU----UCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCCUUAUUCGGGCGCCAA-AGGCUGUUGA -------((((((((((((...----.....))..............((((..((....))((((.....))))))))))))))).))).((((((..-.))).))).. ( -31.30, z-score = -1.65, R) >droSec1.super_1 14145973 92 - 14215200 ----------GGGAAGGACGAU----UCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU--UUGGGGCGCCAA-AGGCUGUUGA ----------.((((((((..(----((.....)))...........((((..((....))((((.....))))))))))))))--))..((((((..-.))).))).. ( -27.20, z-score = -1.26, R) >droSim1.chr2R 15253305 92 - 19596830 -------GGAAAGG---ACGAU----UCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU--UUGGGGCGCCAA-AGGCUGUUGA -------.((((((---((..(----((.....)))...........((((..((....))((((.....))))))))))))))--))..((((((..-.))).))).. ( -27.60, z-score = -1.50, R) >droYak2.chr2R 8537459 97 + 21139217 -----CCAAAAAGGG--ACAAU----UCCCUGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUUCUUUUGGGCGCCAA-AGGCUGUUGA -----((((((((((--((..(----((.....)))...........((((..((....))((((.....)))))))))))))).))))))..(((..-.)))...... ( -29.70, z-score = -1.82, R) >droEre2.scaffold_4845 10742442 101 - 22589142 AAAAACCCAAAAGGG--ACGAU----UCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCCUC-CUCGGGCGCCAA-AGGCUGUUGA .....(((...((((--((..(----((.....)))...........((((..((....))((((.....)))))))))))))).-...))).(((..-.)))...... ( -30.60, z-score = -1.69, R) >droAna3.scaffold_13266 15897301 99 + 19884421 -GAGGCGUUGGGGCGGAGCGGU----CCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGCCCCC--C--AGGCGCCGA-ACGCUGUUGA -..(((((..(((.((.((.((----.....))..............((((..((....))((((.....))))))))))))))--)--..)))))..-.......... ( -37.00, z-score = -1.10, R) >droMoj3.scaffold_6496 25376806 78 + 26866924 ---------------------------CAAGACGAAAUAAAUAAGUAUGACACGCGCGAGCGCAUGAGAAAUGCGGCAGCCCUUU----GGGCCACACAAACGUGUUGA ---------------------------(((.(((..........((.......((....))((((.....)))).)).((((...----))))........))).))). ( -20.70, z-score = -1.13, R) >droGri2.scaffold_15245 15941950 101 - 18325388 ---GGAUGGGCGGUGUUACAAUCUUCCCUUGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGCCCUUU----AGGCAACAC-AACGUGUCGA ---.((((.(..(((((......((((....).)))...........((((..((....))((((.....))))))))(((....----.))))))))-..).)))).. ( -27.90, z-score = -0.29, R) >consensus _______GGAAAGGG__ACGAU____UCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU__UU_GGGCGCCAA_AGGCUGUUGA ..............................(((..............((((..((....))((((.....))))))))((((.......)))))))............. (-19.63 = -20.40 + 0.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:56 2011