
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,444,030 – 3,444,124 |
| Length | 94 |
| Max. P | 0.683996 |

| Location | 3,444,030 – 3,444,124 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.42 |
| Shannon entropy | 0.58690 |
| G+C content | 0.47519 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -10.46 |
| Energy contribution | -10.59 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.683996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
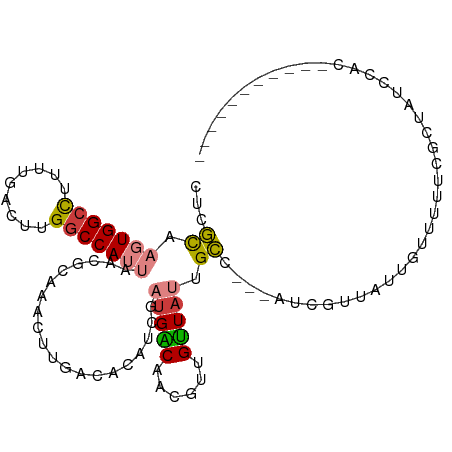
>dm3.chr2L 3444030 94 + 23011544 CUCGCAAGUGGCCUUUUGACUUGGCCAUUAACGCAAACUUGACACAUCGAUGACAACGUUGUUAUAGCC---AUCGUUAUUGUUUUUCGCUAUCCAC----------- ...((.(((((((.........))))))).....((((.((((......((((((....))))))....---...))))..))))...)).......----------- ( -18.42, z-score = -0.42, R) >droSec1.super_129 6115 94 + 51580 GUUGCAAGUGGCCUUUUGACUUGGCCAUUAACGCAAACUUGACACAUCGAUGACAGCGUUGUUAUAGCC---AACGUUAUUGUUUUUCGCUAGCCAC----------- .((((.(((((((.........)))))))...))))...........(((.((((((((((.......)---)))))...))))..)))........----------- ( -22.40, z-score = -0.62, R) >droYak2.chr2L 3431660 93 + 22324452 CUCGCAAGUGGCCUUUUGACUUGGCCAUUAACGCGAACUUGACACAUCGAUGACAACGUGGUUAUUGCC---AACGUUAUUGUUUU-CGCUAUCCAC----------- .((((.(((((((.........)))))))...))))...........(((.(((((((((((....)))---.))))...)))).)-))........----------- ( -26.00, z-score = -2.40, R) >droEre2.scaffold_4929 3476404 93 + 26641161 CUCGCAAGUGGCCUUUCGACUUGGCCAUUAACGCAAACUUGACACAUCGAUGACAACGUUGUUAUUGCC---AUCGUUAUUGUUUU-CGCUAUCCAC----------- ...((.(((((((.........)))))))...(.((((.((((....((((((((....))))))))..---...))))..)))).-))).......----------- ( -22.10, z-score = -1.65, R) >droAna3.scaffold_12916 2907712 108 - 16180835 CGUCUAAGUGGCCUUUUGACUUGGCCAUUAACGUAAACUUGACACAUUGACGGCGAUAUUGUUAUUGGUGGUAUUGCUUUUGGCUUUUGCUUUUUUCCUUUGCACUCG ((((..(((((((.........)))))))...((.......)).....))))(((((((..(.....)..)))))))....(((....)))................. ( -24.30, z-score = -1.12, R) >dp4.chr4_group3 5598721 71 - 11692001 ---GCAAGUGGCUUUUUGACUUGGCCA---------ACUUGACACUUUGAUGGCGG----GCUACGGCU---GGUGUCUUGGCCACAGAC------------------ ---....((((((....(((...((((---------.(..........).)))).(----((....)))---...)))..))))))....------------------ ( -23.60, z-score = -1.51, R) >droPer1.super_1 2695847 71 - 10282868 ---GCAAGUGGGUUUUUGACUUGGCCA---------ACUUGACACUUUGAUGGCGG----GCUACGGCU---GGUGUCUUGGCCACCGAC------------------ ---....((.(((.........(((((---------(...((((((.....(((.(----....).)))---))))))))))))))).))------------------ ( -22.60, z-score = -0.85, R) >consensus CUCGCAAGUGGCCUUUUGACUUGGCCAUUAACGCAAACUUGACACAUCGAUGACAACGUUGUUAUUGCC___AUCGUUAUUGUUUUUCGCUAUCCAC___________ ...((.(((((((.........)))))))....................(((((......))))).))........................................ (-10.46 = -10.59 + 0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:54 2011