
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,545,375 – 16,545,472 |
| Length | 97 |
| Max. P | 0.884213 |

| Location | 16,545,375 – 16,545,472 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.94 |
| Shannon entropy | 0.42218 |
| G+C content | 0.54223 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -26.77 |
| Energy contribution | -26.61 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
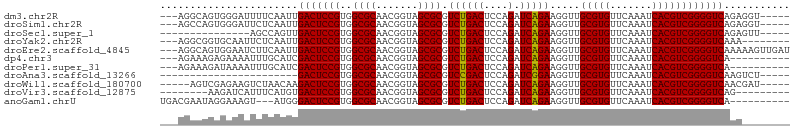
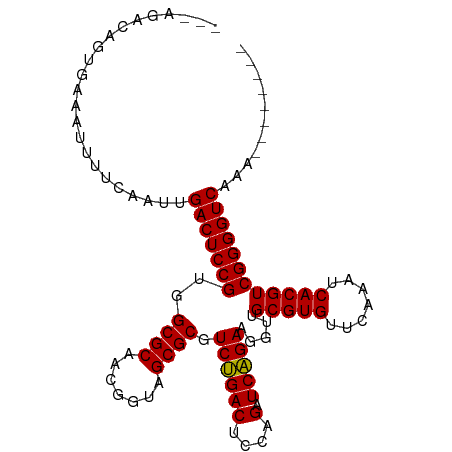
>dm3.chr2R 16545375 97 + 21146708 ---AGGCAGUGGGAUUUUCAAUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAGAGGU----- ---..((..(((..((.......))..)))..))((.......))((.(((((((((.((((....))))(((((.......))))).))))))))).))----- ( -31.50, z-score = -0.85, R) >droSim1.chr2R 15207185 97 + 19596830 ---AGCCAGUGGGAUUCUCAAUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAGAGGU----- ---.((((...(((.((......)).))).))))((.......))((.(((((((((.((((....))))(((((.......))))).))))))))).))----- ( -35.30, z-score = -1.66, R) >droSec1.super_1 14099502 85 + 14215200 ---------------AGCCAGUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAGAGUU----- ---------------.(((.((((.(.(....).))))))))...((.(((((((((.((((....))))(((((.......))))).))))))))))).----- ( -28.40, z-score = -0.88, R) >droYak2.chr2R 8488821 94 - 21139217 ---AGGCGGUGCAAUUCUCAAUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAAA-------- ---..((((.(((((.....)))).).)))).((((.......))))...(((((((.((((....))))(((((.......))))).)))))))..-------- ( -30.60, z-score = -0.93, R) >droEre2.scaffold_4845 10693280 102 + 22589142 ---AGGCAGUGGAAUCUUCAAUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAAAAAGUUGAU ---((((...(((.((.......)).)))...((((.......))))))))((((((.((((....))))(((((.......))))).))))))........... ( -30.20, z-score = -0.63, R) >dp4.chr3 2057438 92 - 19779522 ---AGAAAGAGAAAAUUUGCAUCGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA---------- ---....................(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).---------- ( -26.70, z-score = -0.18, R) >droPer1.super_31 71914 92 + 935084 ---AGAAAGAUAAAAUUUGCAUCGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA---------- ---....................(((((((..((((.......)))).((((((....).))))).....(((((.......)))))))))))).---------- ( -26.70, z-score = -0.52, R) >droAna3.scaffold_13266 12988195 77 - 19884421 -----------------------GACUCCGUGGCGCAACGGUAGCGCGUCCGACUCCAGAUCGGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAAGUCU----- -----------------------((((.....((((.......))))....((((((.((((....))))(((((.......))))).)))))).)))).----- ( -29.50, z-score = -1.71, R) >droWil1.scaffold_180700 1819286 95 - 6630534 -----AGUCGAGAAGUCUAACAAGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAACGAU----- -----.((((.(.(((((....))))).)...((((.......))))...(((((((.((((....))))(((((.......))))).))))))).))))----- ( -30.20, z-score = -0.90, R) >droVir3.scaffold_12875 5376997 88 + 20611582 --------AAGAUCAUUUCAUGUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAG--------- --------...........(((((.((((((......)))).)))))))((((((((.((((....))))(((((.......))))).))))))))--------- ( -29.20, z-score = -1.49, R) >anoGam1.chrU 56108079 92 - 59568033 UGACGAAUAGGAAAGU---AUGGGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCA---------- .(((...........(---((((....)))))((((.......))))))).((((((.((((....))))(((((.......))))).)))))).---------- ( -28.70, z-score = -0.51, R) >consensus ___AGACAGUGAAAUUUUCAAUUGACUCCGUGGCGCAACGGUAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUCAAA________ .......................(((((((..((((.......)))).((((((....).))))).....(((((.......))))))))))))........... (-26.77 = -26.61 + -0.16)
| Location | 16,545,375 – 16,545,472 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.94 |
| Shannon entropy | 0.42218 |
| G+C content | 0.54223 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -22.72 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.86 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
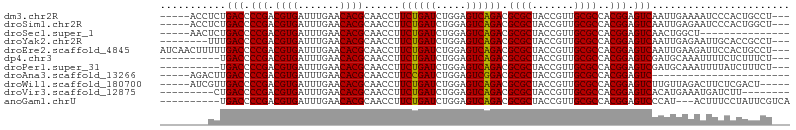
>dm3.chr2R 16545375 97 - 21146708 -----ACCUCUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAAUUGAAAAUCCCACUGCCU--- -----.....((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))...................--- ( -23.50, z-score = -1.18, R) >droSim1.chr2R 15207185 97 - 19596830 -----ACCUCUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAAUUGAGAAUCCCACUGGCU--- -----..(((((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))...))).............--- ( -25.30, z-score = -0.75, R) >droSec1.super_1 14099502 85 - 14215200 -----AACUCUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAACUGGCU--------------- -----.....((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))).......--------------- ( -23.50, z-score = -0.75, R) >droYak2.chr2R 8488821 94 + 21139217 --------UUUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAAUUGAGAAUUGCACCGCCU--- --------.(((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))))..................--- ( -24.30, z-score = -0.83, R) >droEre2.scaffold_4845 10693280 102 - 22589142 AUCAACUUUUUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAAUUGAAGAUUCCACUGCCU--- .....(((((((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))))..))))............--- ( -26.10, z-score = -1.47, R) >dp4.chr3 2057438 92 + 19779522 ----------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCGAUGCAAAUUUUCUCUUUCU--- ----------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))....................--- ( -22.70, z-score = -0.43, R) >droPer1.super_31 71914 92 - 935084 ----------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCGAUGCAAAUUUUAUCUUUCU--- ----------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))((((.......)))).....--- ( -23.00, z-score = -0.75, R) >droAna3.scaffold_13266 12988195 77 + 19884421 -----AGACUUGACCCCGACGUGAUUUGAACACGCAACCUUCCGAUCUGGAGUCGGACGCGCUACCGUUGCGCCACGGAGUC----------------------- -----.(((((....((((((((.......))).......(((.....))))))))..((((.......))))....)))))----------------------- ( -22.80, z-score = -0.67, R) >droWil1.scaffold_180700 1819286 95 + 6630534 -----AUCGUUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCUUGUUAGACUUCUCGACU----- -----...(((((.......(((.................((((((.....)))))).((((.......)))))))(((((((....)))))))))))).----- ( -27.30, z-score = -1.03, R) >droVir3.scaffold_12875 5376997 88 - 20611582 ---------CUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCACAUGAAAUGAUCUU-------- ---------.((((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).))))..............-------- ( -23.20, z-score = -1.06, R) >anoGam1.chrU 56108079 92 + 59568033 ----------UGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCCCAU---ACUUUCCUAUUCGUCA ----------.(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))....---................ ( -21.80, z-score = -0.51, R) >consensus ________UCUGACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUACCGUUGCGCCACGGAGUCAAUUGAAAAUUCCACUGUCU___ ...........(((.(((.((((.......))))......((((((.....)))))).((((.......))))..))).)))....................... (-22.72 = -22.55 + -0.16)
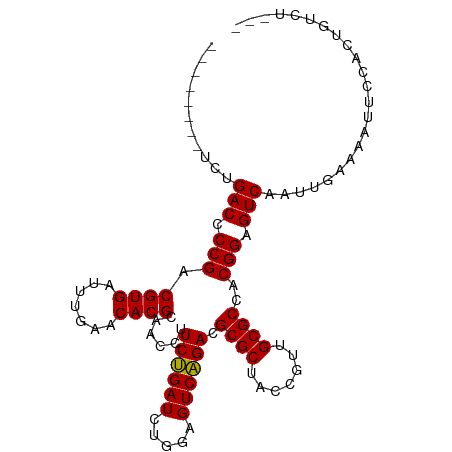
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:52 2011