
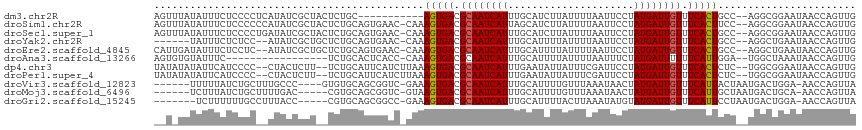
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,504,358 – 16,504,462 |
| Length | 104 |
| Max. P | 0.997334 |

| Location | 16,504,358 – 16,504,462 |
|---|---|
| Length | 104 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.57 |
| Shannon entropy | 0.58883 |
| G+C content | 0.37209 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -11.94 |
| Energy contribution | -11.80 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.997334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
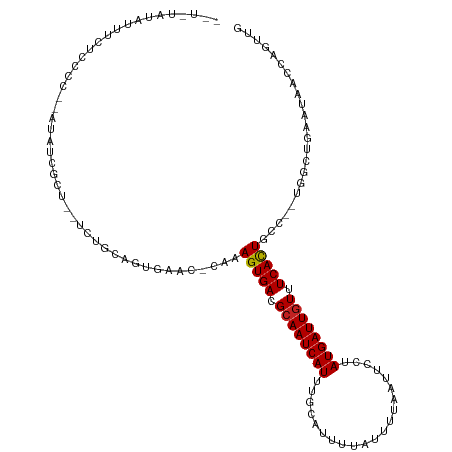
>dm3.chr2R 16504358 104 - 21146708 AGUUUAUAUUUCUCCCCUCAUAUCGCUACUCUGC-----------AGUGACGCAAUCAUUUGCAUCUUAUUUUAAUUCCUAUGAUUGUUUCACUGCC--AGGCGGAAUAACCAGUUG ......................(((((.....((-----------(((((.((((((((.....................)))))))).))))))).--.)))))............ ( -23.90, z-score = -3.27, R) >droSim1.chr2R 15165557 114 - 19596830 AGUUUAUAUUUCUCCCCCCAUAUCGCUACUCUGCAGUGAAC-CAAAGUGACGCAAUCAUUAGCAUCUUAUUUUAAUUCCUAUGAUUGUUUCACUUCC--AGGCGGAAUAACCAGUUG ......((((((.((.......(((((.......)))))..-..((((((.((((((((.....................)))))))).))))))..--.)).))))))........ ( -23.40, z-score = -2.67, R) >droSec1.super_1 14058419 114 - 14215200 AGUUUAUAUUUCUCCCCUGAUAUCGCUACUCUGCAGUGAAC-CAAAGUGACGCAAUCAUUUGCAUCUUAUUUUAAUUCCUAUGAUUGUUUCACUGCC--AGGCGGAAUAACCAGUUG ............(((((((...(((((.......)))))..-...(((((.((((((((.....................)))))))).)))))..)--))).)))........... ( -26.60, z-score = -2.65, R) >droYak2.chr2R 8446809 106 + 21139217 ------UAUUUCUCUCC--AUAUCGCUGCUCUGCAGUGAAC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCC--AGGCUGAAUAACCAGUUG ------...........--...(((((((...)))))))..-...(((((.((((((((..(.(((.......))).)..)))))))).)))))...--.(((((......))))). ( -27.00, z-score = -3.38, R) >droEre2.scaffold_4845 10650764 112 - 22589142 CAUUGAUAUUUCUCCUC--AUAUCGCUGCUCUGCAGUGAAC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCC--AGGCUGAAUAACCAGUUG .((((.(((((..(((.--...(((((((...)))))))..-...(((((.((((((((..(.(((.......))).)..)))))))).)))))...--)))..)))))..)))).. ( -27.90, z-score = -3.02, R) >droAna3.scaffold_13266 15786066 97 + 19884421 AGUGUGUAUUUC-----------------UCUGCACUCACC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUUCUAUGAUUUUUUCAUUGGA--UGGCUAAAUAACCAGUUG ...(((((....-----------------..)))))((((.-....)))).((((....))))...((((((.....(((((((.....))).))))--.....))))))....... ( -15.60, z-score = -0.34, R) >dp4.chr3 11134311 111 + 19779522 UAUAUAUAUUCAUCCCC--CUACUCUU--UCUGCAUUCAUCUUAAAGUGACGCAAUCAUUUGAAUAUUAUUUCGAUUCCUAUGAUUGUUUCACUCUC--UGGCGGAAUAACCAGUUG .................--.......(--(((((...........(((((.((((((((..((((.........))))..)))))))).)))))...--..)))))).......... ( -21.59, z-score = -3.00, R) >droPer1.super_4 4633104 111 + 7162766 UAUAUAUAUUCAUCCCC--CUACUCUU--UCUGCAUUCAUCUUAAAGUGACGCAAUCAUUUGAAUAUUAUUUCGAUUCCUAUGAUUGUUUCACUCUC--UGGCGGAAUAACCAGUUG .................--.......(--(((((...........(((((.((((((((..((((.........))))..)))))))).)))))...--..)))))).......... ( -21.59, z-score = -3.00, R) >droVir3.scaffold_12823 197737 105 + 2474545 ------UUUUUAUCUGCUUUGCCC----GUGUGCAGCGGUC-GAAAGUGACGCAAUCAUUUGCAUUUUGUUUAAAUAACUAUGAUUGUUUCAUUACUAAUGACUGGA-AACCAGUUA ------...........((((.((----((.....)))).)-)))(((((.((((((((....((((.....))))....)))))))).))))).....(((((((.-..))))))) ( -25.70, z-score = -1.94, R) >droMoj3.scaffold_6496 25235110 104 + 26866924 ------UCUUUAUCUGCUUUUGAC-----CGUGCAGCGGUC-GUAAGUGACGCAAUCAUUUGCAUUUUGUUUAAAUAACUAUGAUUGUUUCAUUGCUAAUGACUGCA-AACCAGUUA ------........(((.......-----...)))((((((-((.(((((.((((((((....((((.....))))....)))))))).)))))....)))))))).-......... ( -26.10, z-score = -2.04, R) >droGri2.scaffold_15245 15799040 103 - 18325388 -------UCUUUUUUGCCUUUACC-----CGUGCAGCGGCC-GAAAGUGACGCAAUCAUUUGCAUUUUACUUAAAUAUGUAUGAUUGUUUCAUUCCUAAUGACUGGA-AACCAGUUA -------..........((((..(-----((.....)))..-.))))(((.((((((((..((((.((.....)).)))))))))))).))).......(((((((.-..))))))) ( -22.80, z-score = -1.84, R) >consensus __U_UAUAUUUCUCCCC__AUAUCGCU__UCUGCAGUGAAC_CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCC__UGGCUGAAUAACCAGUUG .............................................(((((.((((((((.....................)))))))).)))))....................... (-11.94 = -11.80 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:48 2011