
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,498,228 – 16,498,319 |
| Length | 91 |
| Max. P | 0.789421 |

| Location | 16,498,228 – 16,498,319 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Shannon entropy | 0.71383 |
| G+C content | 0.45498 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -7.13 |
| Energy contribution | -7.22 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.789421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16498228 91 - 21146708 -----------GUGGGCGUCGCAUA-ACUGUUGGCUUUUCUGCGCCCCGGC-AUUAUUUUAUGGCCAUAUUUAAAAUCGGCUCAAAACCGCUCACGCAAUUCCG -----------(((((((...((((-(......((......))((....))-......)))))(((............))).......)))))))......... ( -21.30, z-score = 0.12, R) >droSim1.chr2R 15159566 91 - 19596830 -----------GUGGGCGUCGCAUA-ACUGUUGGCUUUUCUGCGCCCCGGC-AUUAUUUUAUGGCCAUAUUUAAAAUCCGCUCAAAGCCGCUCACGCAAUUCCG -----------((((((...(((..-..))).((((((...((((....))-...((((((..........))))))..))..))))))))))))......... ( -22.60, z-score = -0.15, R) >droSec1.super_1 14052481 91 - 14215200 -----------GUGGGCGUCGCAUA-ACUGUUGGCUUUUCUGCGCCCCGGC-AUUAUUUUAUGGCCAUAUUUAAAAUCCGCUCAAAACCACUCACGCAAUUCCG -----------.((((((.......-.....(((((.......((....))-..........)))))...........)))))).................... ( -15.13, z-score = 1.37, R) >droYak2.chr2R 8440960 76 + 21139217 -----------GUGGGCGUCGCAUA-ACUGUUGGCUUUUC----------------CUUUAUGGUCAUAUUUAAAAUCCGCUCAAAACCGCUCACGCAAUUCCG -----------(((((((..(((..-..)))(((((....----------------......))))).....................)))))))......... ( -13.70, z-score = -0.44, R) >droEre2.scaffold_4845 10644814 76 - 22589142 -----------GUGGGCGUCGCAUA-ACUGUUGGCUUUUC----------------UUUUAUGGUCAUAUUUAAAAUCCGCUCAAAGCCGCUCACGCAAUUCCG -----------((((((...(((..-..))).((((((..----------------.....(((.............)))...))))))))))))......... ( -15.42, z-score = -0.67, R) >droAna3.scaffold_13266 15780151 90 + 19884421 -----------GUGGGCGUCGCAUA-ACUGUCCGGGUUAU-GCUCUGAGCC-CGAACUUUAUGGUCAUAUUUAAAAUCCGCUCAAAUCCGCUCACGCAAUUCCG -----------(((((((..(((((-(((.....))))))-))..(((((.-........(((....))).........)))))....)))))))......... ( -25.67, z-score = -2.27, R) >dp4.chr3 11128184 92 + 19779522 -----------GUGGGCGUCGCAUA-ACUGUAUGUACGUCUAUGUGCUGCUGCUCGGCUUGUGUUCAUAUUGAAAAUCUACUUUAAGCCGCUCACGCAAUUCCA -----------((((((((.(((((-....)))))))))))))....(((((..(((((((..(((.....))).........)))))))..)).)))...... ( -24.80, z-score = -1.25, R) >droPer1.super_4 4627037 92 + 7162766 -----------GUGGGCGUCGCAUA-ACUGUACGUACGUCUAUGUGCUGCUGCUCGGCUUGUGUUCAUAUUGAAAAUCUACUCUAAGCCGCUCACGCAAUUCCA -----------((((((((.(((..-...(((((((....)))))))))).))..((((((.((...............)).).)))))))))))......... ( -25.06, z-score = -1.28, R) >droWil1.scaffold_180700 3214352 91 + 6630534 CUGAGCACCCUGUGGGCGUC--AUA-ACUGUAGUCGUCUCAG----------UUAUAGUUAUGGUCAUAUUUAAAAUCUACUCUAAGCCGCUCACGCAAUGCCA ..(.(((...((((((((((--(((-(((((((.(......)----------)))))))))))).............((......)).))))))))...)))). ( -23.70, z-score = -2.00, R) >droVir3.scaffold_12823 189625 73 + 2474545 -----------GUGGGCGUCAUAAAUGUUGUUGCUCUCAAUUUUCU---UAUAU-----------UAUA-UUUCAUUU---UUUCGA-CGCUCACGCAAUUCG- -----------(((((((((..(((((..((...............---.....-----------...)-)..)))))---....))-)))))))........- ( -16.50, z-score = -3.98, R) >droMoj3.scaffold_6496 25225024 92 + 26866924 -----------GUGGGCGUCAAAAAUGUUGUUGCUCUCAAUAUACUAUGUAUAUCUGUAUGUGUACAUAUUUUCGUUUUUAUUUUGA-CGCUCACGCAAUUUCA -----------(((((((((((((((((((.......))))))..(((((((((......)))))))))............))))))-)))))))......... ( -30.00, z-score = -6.01, R) >consensus ___________GUGGGCGUCGCAUA_ACUGUUGGCUUUUCUGCGC_C_G_C_AU_AUUUUAUGGUCAUAUUUAAAAUCCGCUCAAAACCGCUCACGCAAUUCCG ...........(((((((......................................................................)))))))......... ( -7.13 = -7.22 + 0.09)
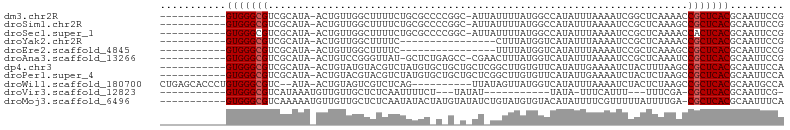

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:47 2011