
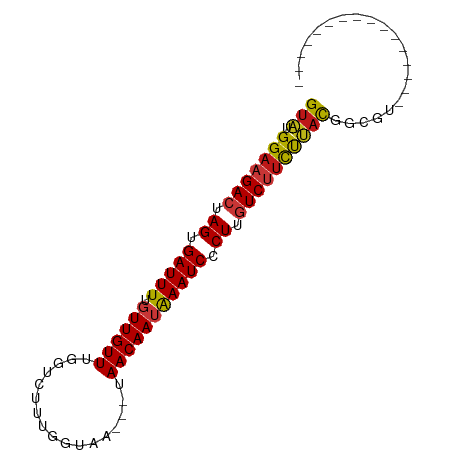
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,493,582 – 16,493,653 |
| Length | 71 |
| Max. P | 0.998495 |

| Location | 16,493,582 – 16,493,653 |
|---|---|
| Length | 71 |
| Sequences | 15 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 84.66 |
| Shannon entropy | 0.30309 |
| G+C content | 0.36400 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -16.70 |
| Energy contribution | -16.69 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.37 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.998495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
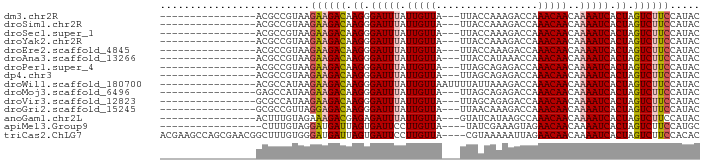
>dm3.chr2R 16493582 71 + 21146708 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.(((((.((((((....(....)...---.))))))))))).)).)))))))))).....---------------- ( -19.20, z-score = -3.04, R) >droSim1.chr2R 15154959 71 + 19596830 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.(((((.((((((....(....)...---.))))))))))).)).)))))))))).....---------------- ( -19.20, z-score = -3.04, R) >droSec1.super_1 14047884 71 + 14215200 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.(((((.((((((....(....)...---.))))))))))).)).)))))))))).....---------------- ( -19.20, z-score = -3.04, R) >droYak2.chr2R 8436399 71 - 21139217 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.(((((.((((((....(....)...---.))))))))))).)).)))))))))).....---------------- ( -19.20, z-score = -3.04, R) >droEre2.scaffold_4845 10640199 71 + 22589142 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.(((((.((((((....(....)...---.))))))))))).)).)))))))))).....---------------- ( -19.20, z-score = -3.04, R) >droAna3.scaffold_13266 15774978 71 - 19884421 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUUUAUGGUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.((((((((((((...........))---)))))..))))).)).)))))))))).....---------------- ( -18.80, z-score = -2.93, R) >droPer1.super_4 4622595 71 - 7162766 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUCUGCUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.((((((((((((.(((....)))))---)))))..))))).)).)))))))))).....---------------- ( -20.80, z-score = -3.99, R) >dp4.chr3 11123787 71 - 19779522 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUCUGCUAA---UAACAAUAAAUCCCUUGUCUUCUUACGGCGU---------------- (((.(((((((.((.((((((((((((.(((....)))))---)))))..))))).)).)))))))))).....---------------- ( -20.80, z-score = -3.99, R) >droWil1.scaffold_180700 3206868 74 - 6630534 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUAAUAAAAUUAACAAUAAAUCCCUUGUCUUCUUAUGGCGU---------------- ((..(((((((.((.(((((.((((((.................))))))))))).)).)))))))....))..---------------- ( -17.93, z-score = -3.33, R) >droMoj3.scaffold_6496 25216804 71 - 26866924 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUCUGCUAA---UAACAAUAAAUCCCUUGUCUUCUUAUGGCUC---------------- ((..(((((((.((.((((((((((((.(((....)))))---)))))..))))).)).)))))))....))..---------------- ( -19.90, z-score = -4.22, R) >droVir3.scaffold_12823 179291 71 - 2474545 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUCUGCUAA---UAACAAUAAAUCCCUUGUCUUCUUAUGGCGC---------------- ((..(((((((.((.((((((((((((.(((....)))))---)))))..))))).)).)))))))....))..---------------- ( -20.40, z-score = -4.01, R) >droGri2.scaffold_15245 15782227 71 + 18325388 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGUUAA---UAACAAUAAAUCCCUUGUCUCCUAACGGCGC---------------- ((..(((.(((.((.((((((((((((...........))---)))))..))))).)).))))))..)).....---------------- ( -15.00, z-score = -1.58, R) >anoGam1.chr2L 41010212 71 - 48795086 GUAUGGAAGACUAGUGAUUUUGUUGUUUGGCUUAUGAUAC---UAACAAUAAAUCUCUCGUCUUUCUACAAAGU---------------- (((.(((((((.((.(((((.((((((.(..........)---.))))))))))).)).)))))))))).....---------------- ( -20.00, z-score = -3.88, R) >apiMel3.Group9 6557012 69 - 10282195 GCAUGGAAGACUAGUGAUUUUGUUGUUCUACUUUCGAUA----UAACAAGGAAUCACUAAUCAUCCUACAAAG----------------- ....(((.((.(((((((((((((((((.......)).)----)))))..))))))))).)).))).......----------------- ( -18.70, z-score = -3.73, R) >triCas2.ChLG7 2378810 86 + 17478683 GUGUGGAAGACUAGUGAUUUUGUUGUUCUAAUUUUUACG----UAACAAGGAAUCACUAAUCAUCCCACAAAGCCGUUCGCUGGCUUCGU (((.(((.((.((((((((((.((((((..........)----.))))))))))))))).)).)))))).((((((.....))))))... ( -26.80, z-score = -3.69, R) >consensus GUAUGGAAGACUAGUGAUUUUGUUGUUUGGUCUUUGGUAA___UAACAAUAAAUCCCUUGUCUUCUUACGGCGU________________ (((.(((((((.((.(((((.((((((.................))))))))))).)).))))))))))..................... (-16.70 = -16.69 + -0.01)

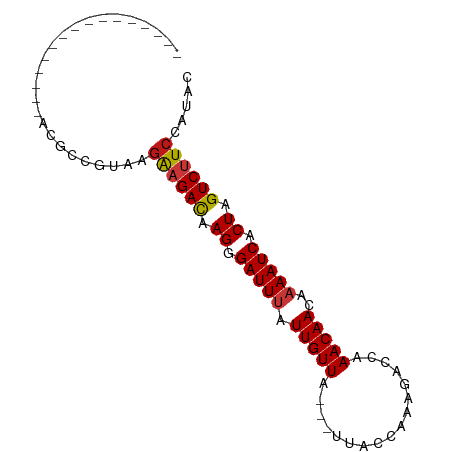
| Location | 16,493,582 – 16,493,653 |
|---|---|
| Length | 71 |
| Sequences | 15 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 84.66 |
| Shannon entropy | 0.30309 |
| G+C content | 0.36400 |
| Mean single sequence MFE | -14.77 |
| Consensus MFE | -11.41 |
| Energy contribution | -11.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16493582 71 - 21146708 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.60, R) >droSim1.chr2R 15154959 71 - 19596830 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.60, R) >droSec1.super_1 14047884 71 - 14215200 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.60, R) >droYak2.chr2R 8436399 71 + 21139217 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.60, R) >droEre2.scaffold_4845 10640199 71 - 22589142 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.60, R) >droAna3.scaffold_13266 15774978 71 + 19884421 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUACCAUAAACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.92, R) >droPer1.super_4 4622595 71 + 7162766 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUAGCAGAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.28, R) >dp4.chr3 11123787 71 + 19779522 ----------------ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA---UUAGCAGAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.28, R) >droWil1.scaffold_180700 3206868 74 + 6630534 ----------------ACGCCAUAAGAAGACAAGGGAUUUAUUGUUAAUUUUAUUAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((..((((....))))...)))))..))))).)).))))))..... ( -13.40, z-score = -3.28, R) >droMoj3.scaffold_6496 25216804 71 + 26866924 ----------------GAGCCAUAAGAAGACAAGGGAUUUAUUGUUA---UUAGCAGAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.70, R) >droVir3.scaffold_12823 179291 71 + 2474545 ----------------GCGCCAUAAGAAGACAAGGGAUUUAUUGUUA---UUAGCAGAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -13.04, z-score = -2.65, R) >droGri2.scaffold_15245 15782227 71 - 18325388 ----------------GCGCCGUUAGGAGACAAGGGAUUUAUUGUUA---UUAACAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.........((((((.((.(((((.(((((.---.............)))))..))))).)).))))))..... ( -12.54, z-score = -1.88, R) >anoGam1.chr2L 41010212 71 + 48795086 ----------------ACUUUGUAGAAAGACGAGAGAUUUAUUGUUA---GUAUCAUAAGCCAAACAACAAAAUCACUAGUCUUCCAUAC ----------------.....(((..(((((.((.(((((.(((((.---((.......))..)))))..))))).)).)))))...))) ( -11.10, z-score = -1.57, R) >apiMel3.Group9 6557012 69 + 10282195 -----------------CUUUGUAGGAUGAUUAGUGAUUCCUUGUUA----UAUCGAAAGUAGAACAACAAAAUCACUAGUCUUCCAUGC -----------------.......(((.(((((((((((..(((((.----...(....)...)))))...))))))))))).))).... ( -23.40, z-score = -5.26, R) >triCas2.ChLG7 2378810 86 - 17478683 ACGAAGCCAGCGAACGGCUUUGUGGGAUGAUUAGUGAUUCCUUGUUA----CGUAAAAAUUAGAACAACAAAAUCACUAGUCUUCCACAC ..((((((.......))))))((((((.(((((((((((..(((((.----(..........))))))...))))))))))))))))).. ( -30.70, z-score = -5.46, R) >consensus ________________ACGCCGUAAGAAGACAAGGGAUUUAUUGUUA___UUACCAAAGACCAAACAACAAAAUCACUAGUCUUCCAUAC .........................((((((.((.(((((.(((((.................)))))..))))).)).))))))..... (-11.41 = -11.56 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:47 2011