| Sequence ID | dm3.chr2R |
|---|---|
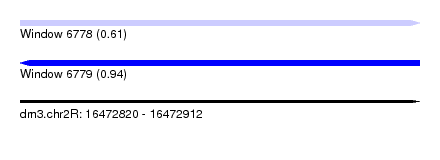
| Location | 16,472,820 – 16,472,912 |
| Length | 92 |
| Max. P | 0.939943 |

| Location | 16,472,820 – 16,472,912 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 53.81 |
| Shannon entropy | 0.84586 |
| G+C content | 0.35453 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -1.86 |
| Energy contribution | -2.80 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.10 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16472820 92 + 21146708 -------------UAACUUUCUCGAAGUACUUAGUACCAGAAACGUGUACUUUACAUAAACAUAAAAAAUAACACUUUGCUGAUACCA-GGCACUUGGGAAAAUCA -------------....((((((.((((.(((.(((.(((....((((.......................))))....))).))).)-)).)))))))))).... ( -17.20, z-score = -2.00, R) >droSim1.chr2R 15134514 86 + 19596830 -------------UAAGUUUCUCAAAGGGCUUGGUUCCAAAAAAGGGUACUAUACAA---CA---AAAAUUACACCUCUCUGAAACCA-GGCACUUGCGAAAAUCA -------------....((((.(((...((((((((.((....((((((........---..---.....))).)))...)).)))))-)))..))).)))).... ( -18.96, z-score = -2.17, R) >droSec1.super_1 14027539 89 + 14215200 -------------UAAGUUUCUCAAAGGGCUUGGUUCCAGAAACGUGUACUAUACAAACACA---AAAAUUACACCUAUCUGAUACCA-GGCACUUCCGAAAAUCA -------------....((((...(((.(((((((..((((...(((((.............---.....)))))...))))..))))-))).)))..)))).... ( -21.77, z-score = -3.59, R) >droYak2.chr2R 8415457 89 - 21139217 -------------UAGGUUUCUCGAAGUACGUAGUACCAGUAACGUGUACUUUAACCAAACA---AAAAUUACACCUUGCUGAUACUA-AGCACUUGGGAAAAUCA -------------..(((((..(.((((.(.(((((.((((((.(((((.(((.........---.))).))))).)))))).)))))-.).)))).)..))))). ( -26.90, z-score = -4.19, R) >droEre2.scaffold_4845 10619162 89 + 22589142 -------------CAUGUUUCUCAAAGUACCUAGUACCAGUAACGUGUACUUUACCCAAAUA---AAAAUUACACCUUGCUGAUACUA-UGCACUUGGGAAAAUCA -------------....((((((((.((.(.(((((.((((((.(((((.(((.........---.))).))))).)))))).)))))-.).)))))))))).... ( -24.80, z-score = -4.55, R) >dp4.chr3 11100407 101 - 19779522 UGAAAACAUGAAACGAGCGUUUGGGAACCGAUUACAUUUGAAUUUAACCGUAUGUACA-AAAUGCAUACAUACAUAGUUUAAGCGACGCGACGCGAGAAAGA---- .............((.(((((...(((((((......))).........(((((((..-...))))))).......))))....)))))....)).......---- ( -16.50, z-score = -0.14, R) >triCas2.chrUn_95 45657 80 + 72281 -------------CAAGUUUUUUUUAAUUUUUAACAAAAAGCCCAAAUACUAAUUUU---CA----AGGACAUAAGUU-----CAAUG-GACGUUUAAAAAUGUCG -------------...((((((((((.....))).)))))))...............---..----.((((....)))-----)....-((((((....)))))). ( -9.20, z-score = -0.36, R) >consensus _____________UAAGUUUCUCAAAGUACUUAGUACCAGAAACGUGUACUAUACACA_ACA___AAAAUUACACCUUGCUGAUACCA_GGCACUUGGGAAAAUCA .................((((((((......((((..((((.....................................))))..))))......)))))))).... ( -1.86 = -2.80 + 0.95)

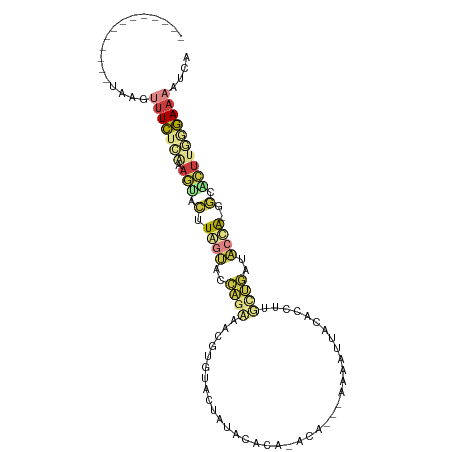
| Location | 16,472,820 – 16,472,912 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 53.81 |
| Shannon entropy | 0.84586 |
| G+C content | 0.35453 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -3.12 |
| Energy contribution | -3.66 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.84 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.13 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.939943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16472820 92 - 21146708 UGAUUUUCCCAAGUGCC-UGGUAUCAGCAAAGUGUUAUUUUUUAUGUUUAUGUAAAGUACACGUUUCUGGUACUAAGUACUUCGAGAAAGUUA------------- (((((((((.((((((.-(((((((((....((((....((((((......)))))).))))....))))))))).)))))).).))))))))------------- ( -30.10, z-score = -5.36, R) >droSim1.chr2R 15134514 86 - 19596830 UGAUUUUCGCAAGUGCC-UGGUUUCAGAGAGGUGUAAUUUU---UG---UUGUAUAGUACCCUUUUUUGGAACCAAGCCCUUUGAGAAACUUA------------- ...((((((.(((.((.-((((((((((((((.(((.....---..---........))).)))))))))))))).)).))))))))).....------------- ( -25.76, z-score = -3.50, R) >droSec1.super_1 14027539 89 - 14215200 UGAUUUUCGGAAGUGCC-UGGUAUCAGAUAGGUGUAAUUUU---UGUGUUUGUAUAGUACACGUUUCUGGAACCAAGCCCUUUGAGAAACUUA------------- ...((((((((.(.((.-((((.(((((.(.(((((....(---((((....)))))))))).).))))).)))).))).)))))))).....------------- ( -25.20, z-score = -2.65, R) >droYak2.chr2R 8415457 89 + 21139217 UGAUUUUCCCAAGUGCU-UAGUAUCAGCAAGGUGUAAUUUU---UGUUUGGUUAAAGUACACGUUACUGGUACUACGUACUUCGAGAAACCUA------------- ....(((((.((((((.-(((((((((.((.(((((...((---((......)))).))))).)).))))))))).)))))).).))))....------------- ( -28.50, z-score = -3.79, R) >droEre2.scaffold_4845 10619162 89 - 22589142 UGAUUUUCCCAAGUGCA-UAGUAUCAGCAAGGUGUAAUUUU---UAUUUGGGUAAAGUACACGUUACUGGUACUAGGUACUUUGAGAAACAUG------------- ((.(((((..((((((.-(((((((((.((.(((((...((---(((....))))).))))).)).))))))))).)))))).))))).))..------------- ( -30.00, z-score = -4.45, R) >dp4.chr3 11100407 101 + 19779522 ----UCUUUCUCGCGUCGCGUCGCUUAAACUAUGUAUGUAUGCAUUU-UGUACAUACGGUUAAAUUCAAAUGUAAUCGGUUCCCAAACGCUCGUUUCAUGUUUUCA ----........(((..((((......((((..((((((((......-.))))))))(((((.((....)).))))))))).....)))).)))............ ( -17.00, z-score = -0.75, R) >triCas2.chrUn_95 45657 80 - 72281 CGACAUUUUUAAACGUC-CAUUG-----AACUUAUGUCCU----UG---AAAAUUAGUAUUUGGGCUUUUUGUUAAAAAUUAAAAAAAACUUG------------- ....((((((((..(((-((...-----.(((.((.....----..---...)).)))...)))))......)))))))).............------------- ( -7.40, z-score = 0.75, R) >consensus UGAUUUUCCCAAGUGCC_UGGUAUCAGAAAGGUGUAAUUUU___UGU_UGUGUAUAGUACACGUUUCUGGUACUAAGUACUUCGAGAAACUUA_____________ ....((((..........(((((((((.((.(.(((.....................))).).)).)))))))))..........))))................. ( -3.12 = -3.66 + 0.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:45 2011