| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,457,690 – 16,457,781 |
| Length | 91 |
| Max. P | 0.782742 |

| Location | 16,457,690 – 16,457,781 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 83.01 |
| Shannon entropy | 0.36221 |
| G+C content | 0.40686 |
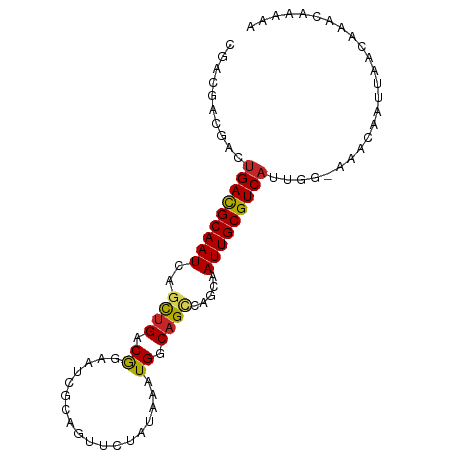
| Mean single sequence MFE | -19.40 |
| Consensus MFE | -12.97 |
| Energy contribution | -12.82 |
| Covariance contribution | -0.15 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.782742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
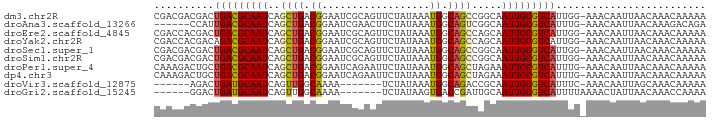
>dm3.chr2R 16457690 91 - 21146708 CGACGACGACUGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCGGCAAUUGCGUCAUUGG-AAACAAUUAACAAACAAAAA ..........(((((((((.(((((.((....))))))).........((.....)).)))))))))(((.-...))).............. ( -21.00, z-score = -1.22, R) >droAna3.scaffold_13266 15737235 85 + 19884421 ------CCAUUGACGCAAUCAGCUGACGGAAUCGAACUUCUAUAAAUGGCAGUCGGCAAUUGCGUCAUUUG-AAACAAUUAACAAAGACAGA ------.((.(((((((((..((((((((((......))))..........)))))).)))))))))..))-.................... ( -20.90, z-score = -2.15, R) >droEre2.scaffold_4845 10601772 91 - 22589142 CGACCACGACUGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCAGCAAUUGCGUCAUUGG-AAACAAUUAACAAACAAAAA ..........(((((((((.(((((.((....))))))).........((.....)).)))))))))(((.-...))).............. ( -21.10, z-score = -1.73, R) >droYak2.chr2R 8400516 91 + 21139217 CGACCACGACAGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCAGCAAUUGCGUCAUUGG-AAACAAUUAACAAACAAAAA ...........((((((((.(((((.((....))))))).........((.....)).))))))))((((.-...))))............. ( -21.30, z-score = -2.12, R) >droSec1.super_1 14012064 91 - 14215200 CGACGACGACUGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCGGCAAUUGCGUCAUUGG-AAACAAUUAACAAACAAAAA ..........(((((((((.(((((.((....))))))).........((.....)).)))))))))(((.-...))).............. ( -21.00, z-score = -1.22, R) >droSim1.chr2R 15119124 91 - 19596830 CGACGACGACUGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCGGCAAUUGCGUCAUUGG-AAACAAUUAACAAACAAAAA ..........(((((((((.(((((.((....))))))).........((.....)).)))))))))(((.-...))).............. ( -21.00, z-score = -1.22, R) >droPer1.super_4 4580820 91 + 7162766 CAAAGACUGCUGACGCAAUCAGCUGACGGAAUCAGAAUUCUAUAAAUGGCAGCUAGAAAUUGCGUCAUUUG-AAACAAUUAACAAACAAAAA ..........(((((((((((((((.((.....((....)).....)).))))).))..))))))))....-.................... ( -19.10, z-score = -2.17, R) >dp4.chr3 11084409 91 + 19779522 CAAAGACUGCUGACGCAAUCAGCUGACGGAAUCAGAAUUCUAUAAAUGGCAGCUAGAAAUUGCGUCAUUUG-AAACAAUUAACAAACAAAAA ..........(((((((((((((((.((.....((....)).....)).))))).))..))))))))....-.................... ( -19.10, z-score = -2.17, R) >droVir3.scaffold_12875 13651679 78 - 20611582 ------AGACUGAUGCAAUCAGUUGGCAAAA-------UCUAUAAAUGGCAGACCGCAAUUGCGUCAUUUC-AAACAAUUAGCAAACAAAAA ------.((.((((((((((.(((.((...(-------(......)).)).))).)..)))))))))..))-.................... ( -12.20, z-score = -0.04, R) >droGri2.scaffold_15245 744201 79 + 18325388 ------GGACUGAUGCAAUCAGUUGGCAAAA-------UCUAUAAGUGACCGAUUGCAAUUGCGUCAUUUUAAAACUAUUAACAAACCAAAA ------(((.((((((((((((((((((...-------........)).)))))))..))))))))).)))..................... ( -17.30, z-score = -2.47, R) >consensus CGACGACGACUGACGCAAUCAGCUGACGGAAUCGCAGUUCUAUAAAUGGCAGCCAGCAAUUGCGUCAUUGG_AAACAAUUAACAAACAAAAA ..........(((((((((..((((.((..................)).)))).....)))))))))......................... (-12.97 = -12.82 + -0.15)
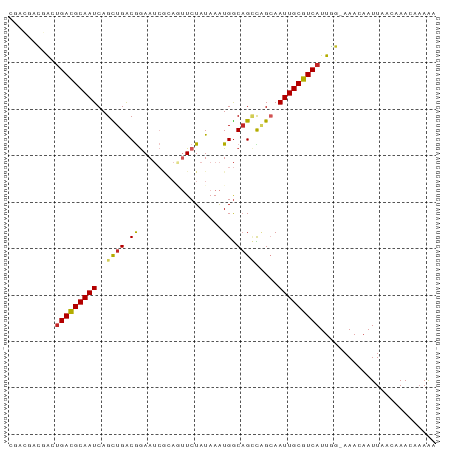
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:40 2011