| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,456,157 – 16,456,259 |
| Length | 102 |
| Max. P | 0.986927 |

| Location | 16,456,157 – 16,456,259 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 81.92 |
| Shannon entropy | 0.33468 |
| G+C content | 0.51919 |
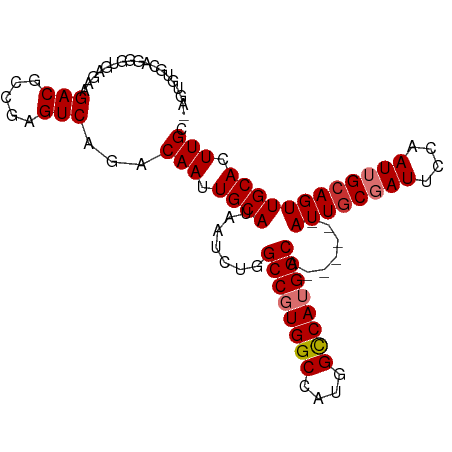
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -28.56 |
| Energy contribution | -29.07 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.986927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
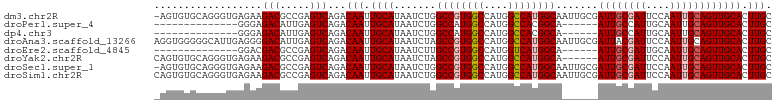
>dm3.chr2R 16456157 102 - 21146708 -AGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAAUUGCAGUUGCACUUGC -...((((((((((......))))(((((.(.((((((((.......((((((((....))))))))...)))))))))))))).........)))))).... ( -42.70, z-score = -2.91, R) >droPer1.super_4 4579588 83 + 7162766 --------------GGGAGACAUUGAGUCAGACAAUUGCAUAAUCUGGCCAUGGCCAUGGCCACGGCA------AUUGCCAUUGCAAUUGCAGUUGCACUUGC --------------.(....)...(((((((.((((((((.....(((((((....))))))).(((.------...)))..))))))))...))).)))).. ( -30.70, z-score = -1.85, R) >dp4.chr3 11083193 83 + 19779522 --------------GGGAGACAUUGAGUCAGACAAUUGCAUAAUCUGGCCAUGGCCAUGGCCACGGCA------AUUGCCAUUGCAAUUGCAGUUGCACUUGC --------------.(....)...(((((((.((((((((.....(((((((....))))))).(((.------...)))..))))))))...))).)))).. ( -30.70, z-score = -1.85, R) >droAna3.scaffold_13266 15735954 103 + 19884421 AGGUGGGGGCAUUGAGGGGACAUUGAGUCAGACAAUUGCAUAAUCUAGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUACGAUUCCAAUUGCAGUUGCACUUGC (((((..(((.(..((....).)..))))...(((((((........((((((((....))))))))(((((.((.......))))))))))))))))))).. ( -37.70, z-score = -2.19, R) >droEre2.scaffold_4845 10600271 83 - 22589142 --------------GGACGACGCCGAGUCAGACAAUUGCAUAAUCUUGCCGUGGCCAUGGUCAUGGCA------AUUGCGAUUGCAAUUGCAGUUGCACUUGC --------------....(((.....)))........(((.....((((((((((....)))))))))------).(((((((((....)))))))))..))) ( -32.60, z-score = -2.50, R) >droYak2.chr2R 8398940 97 + 21139217 CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUAGCCGUGGCCAUGGCCAUGGCA------AUUGCGAUUCCAAUUGCAGUUGCACUUGC ......(((((.......(((.....))).......(((........((((((((....))))))))(------((((((((....))))))))))))))))) ( -36.20, z-score = -2.02, R) >droSec1.super_1 14010577 102 - 14215200 -AGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAAUUGCAGUUGCACUUGC -...((((((((((......))))(((((.(.((((((((.......((((((((....))))))))...)))))))))))))).........)))))).... ( -42.70, z-score = -2.91, R) >droSim1.chr2R 15117604 103 - 19596830 CAGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCAAUUGCGAUUGCGAUUCCAAUUGCAGUUGCACUUGC ((((((((((((((......))))..(.....)..)))))))..)))((((((((....))))))))...((((((((((((....))))))))))))..... ( -43.30, z-score = -2.94, R) >consensus _AGUGUGCAGGGUGAGAAGACGCCGAGUCAGACAAUUGCAUAAUCUGGCCGUGGCCAUGGCCAUGGCA______AUUGCGAUUCCAAUUGCAGUUGCACUUGC ..................(((.....)))...(((.((((.......((((((((....)))))))).......((((((((....)))))))))))).))). (-28.56 = -29.07 + 0.52)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:39 2011