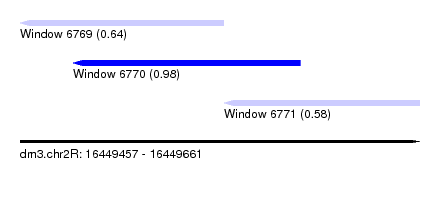
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,449,457 – 16,449,661 |
| Length | 204 |
| Max. P | 0.983714 |

| Location | 16,449,457 – 16,449,561 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.66 |
| Shannon entropy | 0.32636 |
| G+C content | 0.61157 |
| Mean single sequence MFE | -45.57 |
| Consensus MFE | -29.95 |
| Energy contribution | -30.23 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.636842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16449457 104 - 21146708 UGGCGACCGCUGGUGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUGG------GCCCGUGCCCGAUGCUGGACACGCUU .(((....)))(((((((...(((((((..((....))..)))))))....(((((((((((....)))))....(((------((....))))).))))))))))))). ( -45.40, z-score = -1.76, R) >droSim1.chr2R 15110811 104 - 19596830 UGGCGGCCGCUGGUGUGUGACGUGUGAUCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUGG------GCCCGUGCCCGAUGCUGGACACGCUU .(((((((..((((((.((..(((((((..((....))..)))))))........)).)))))).))).(((((((((------((....)))))..))))))..)))). ( -44.30, z-score = -0.98, R) >droSec1.super_1 14003916 104 - 14215200 UGGCGGCCGCUGGUGUGUGACGUGUGAUUGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUGG------GCCCGUGCCCGAUGCUGGACACGCUU .(((((((..((((((.((..(((((((((((....)))))))))))........)).)))))).))).(((((((((------((....)))))..))))))..)))). ( -46.90, z-score = -2.00, R) >droYak2.chr2R 8392093 110 + 21139217 UGGCGGUCGCUGGAGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCAGCUGGGCCCGUGCCCGUGCCCGAUGCUGGACACGCUU .(((((((((((.(((((...(((((((..((....))..)))))))))))))))).(((((....)))))(((((((((.((....)).)))))..))))))).)))). ( -49.70, z-score = -2.09, R) >droEre2.scaffold_4845 10593640 110 - 22589142 UGGCGGUCGCUGGAGUGUGACGUGUGACCAGCGACUGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGUAUCCAGCUGGGCCCGUGCCCGUGCCCGAUGCUGGACACGCUU .(((((((((......)))))(((((((.(((....))).)))))))............))))(((((.(((((((((((.((....)).)))))..)))))).)).))) ( -47.70, z-score = -2.14, R) >droAna3.scaffold_13266 15728781 109 + 19884421 UGUCGCGCUUUGUUGACUUUGCUGACUGCGUCUGCAGCU-GUGACAACACUACGGUAGAUGCGGCCAACUCCAGCUGGGCCCCUGCCCGUGCCCGAUGCUGGACACGCUC ....(((....((((.(......).((((((((((....-(((....)))....)))))))))).))))(((((((((((.(......).)))))..))))))..))).. ( -39.40, z-score = -1.08, R) >consensus UGGCGGCCGCUGGUGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCAGCUGG______GCCCGUGCCCGAUGCUGGACACGCUU .(((...(((....)))....(((((((.(((....))).))))))).....((((.(((((....)))))..)))).......)))((((.(((....))).))))... (-29.95 = -30.23 + 0.28)

| Location | 16,449,484 – 16,449,600 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.07 |
| Shannon entropy | 0.43241 |
| G+C content | 0.54064 |
| Mean single sequence MFE | -41.42 |
| Consensus MFE | -28.25 |
| Energy contribution | -30.50 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.983714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16449484 116 - 21146708 AAAUCCCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCUUGGCGACCGCUGGUGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUG ...((((((.((.....)).)))).)).....((((((..(....).)))))).......(((((((..((....))..))))))).....((((.(((((....)))))..)))) ( -42.10, z-score = -1.83, R) >droSim1.chr2R 15110838 115 - 19596830 -AAAUCCGAAUAUCCGAGAAUCGGCGAAAUCUGCCAGCUUGGCGGCCGCUGGUGUGUGACGUGUGAUCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUG -......((((((.((...(((((((....(((((.....))))).)))))))...))..(((((((..((....))..)))))))))))))(((.(((((....)))))..))). ( -43.60, z-score = -1.87, R) >droSec1.super_1 14003943 115 - 14215200 -AAAUCCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCUUGGCGGCCGCUGGUGUGUGACGUGUGAUUGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCGGCUG -............(((((....((((.....))))..)))))((((((((((.((((...(((((((((((....)))))))))))))))))))).(((((....))))).))))) ( -44.70, z-score = -2.24, R) >droYak2.chr2R 8392126 116 + 21139217 AAAAUCCAAAUUCCCGAGAAUUGGCGAAAUCUGCCAGCUUGGCGGUCGCUGGAGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCAGCUG ............((((((...(((((.....))))).))))).)(((((......)))))(((((((..((....))..))))))).....((((.(((((....)))))..)))) ( -43.60, z-score = -2.87, R) >droEre2.scaffold_4845 10593673 116 - 22589142 AGAAUCCGAAUUGCCGAGAAUCGGCGAAAUCUGCCAGCUUGGCGGUCGCUGGAGUGUGACGUGUGACCAGCGACUGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGUAUCCAGCUG .......((.((((((((....((((.....))))..))))))))))((((((((((...(((((((.(((....))).)))))))))))))....(((((....))))))))).. ( -43.90, z-score = -2.54, R) >droAna3.scaffold_13266 15728814 98 + 19884421 -----------------AAAUAUCUGAGAUUUGCCAGCUUGUCGCGCUUUGUUGACUUUGCUGACUGCGUCUGCAGCU-GUGACAACACUACGGUAGAUGCGGCCAACUCCAGCUG -----------------......(((.((.(((.((((..((((........))))...)))).((((((((((....-(((....)))....)))))))))).))).)))))... ( -30.60, z-score = -1.22, R) >consensus _AAAUCCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCUUGGCGGCCGCUGGUGUGUGACGUGUGACCGGCGACAGCUAGUCAUACAUAUUCAGUAGAUGCCGAGGCAUCCAGCUG ..............((...(((((((....(((((.....))))).)))))))...))..(((((((.(((....))).))))))).....((((.(((((....)))))..)))) (-28.25 = -30.50 + 2.25)

| Location | 16,449,561 – 16,449,661 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 82.14 |
| Shannon entropy | 0.29448 |
| G+C content | 0.47670 |
| Mean single sequence MFE | -23.26 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.580196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16449561 100 - 21146708 AGCCGCCGAACGUGUAAAACAUUCACAAUUCGAUUUGCACACACAGAUUCGAUCCGAAU--CCAAAU-----------------CCCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCU ...((((((..(((((((.(...........).))))))).....((((((...)))))--)....(-----------------(.((......)).)).))))))............. ( -22.00, z-score = -1.69, R) >droSim1.chr2R 15110915 99 - 19596830 AGCCGCUGAACGUGUGAAACAUCCACAAUUCGAUUUGCACACACAGAUUCGAUCCGAAU--CCAAAU------------------CCGAAUAUCCGAGAAUCGGCGAAAUCUGCCAGCU ....((((...(((((....(((........)))...))))).((((((((..((((.(--(....(------------------(.(.....).)))).)))))).)))))).)))). ( -23.90, z-score = -1.61, R) >droSec1.super_1 14004020 99 - 14215200 AGCCGCUGAACGUGUGAAACAUCCACAAUUCGAUUUGCACACACAGAUUCGAUCCGAAC--CCAAAU------------------CCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCU ....((((...(((((....(((........)))...))))).((((((((....(...--.)....------------------((((.((.....)).)))))).)))))).)))). ( -22.80, z-score = -1.71, R) >droYak2.chr2R 8392203 115 + 21139217 AUCCGUUGAGCGUGUGACACAUCCACAAUUCGAUUAGCACACACAGAUUCGUUCCGAAUCCCCGAAUCCAA----AAUCUAAAAUCCAAAUUCCCGAGAAUUGGCGAAAUCUGCCAGCU ....((((.(((((((....(((........)))...)))))...((((((...........))))))...----...............(((((((...)))).)))....)))))). ( -21.20, z-score = -0.90, R) >droEre2.scaffold_4845 10593750 117 - 22589142 AUCCGUUGAGCGUGUAACACAUCCGCAAUUCGAUUUGCACACACAGAUUCGAUCCGAUU--CCAAAUCCAAAUCCAAAUCAGAAUCCGAAUUGCCGAGAAUCGGCGAAAUCUGCCAGCU ....((((.((((((((...(((........)))))))))....(((((...((.((((--(...................))))).)).((((((.....))))))))))))))))). ( -26.41, z-score = -1.18, R) >consensus AGCCGCUGAACGUGUGAAACAUCCACAAUUCGAUUUGCACACACAGAUUCGAUCCGAAU__CCAAAU__________________CCGAAUCCCCGAGAAUCGGCGAAAUCUGCCAGCU ....((((...(((((....(((........)))...))))).((((((((....(......)......................((((.((.....)).)))))).)))))).)))). (-17.48 = -17.40 + -0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:39 2011