
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,449,205 – 16,449,309 |
| Length | 104 |
| Max. P | 0.736324 |

| Location | 16,449,205 – 16,449,309 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 64.73 |
| Shannon entropy | 0.68945 |
| G+C content | 0.50293 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -12.46 |
| Energy contribution | -11.27 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.70 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.736324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16449205 104 - 21146708 ---CAGCACCCCCACCCACCAAUUU---------CCCCCCAAAAAGAUGCGAACGCGCAAGGUGC---UAUUGGUAAUUGGCUUUCUGGUUACGUGGACCUAUGCCACCUACUACCUGC ---.((((((...............---------.....(.....).((((....)))).)))))---)..(((((..((((.....((((.....))))...))))..)))))..... ( -24.70, z-score = -0.51, R) >droSim1.chr2R 15110547 104 - 19596830 ---CAGCACCCACACCCACCAAUUU---------CCCCCCAAGAAGAUGCGAACGCGCAAGGUGC---UGUUGGUAAUUGGCUUUCUGGUUACGUGGACCUAUGCCACCUACUACCUGC ---(((((((............(((---------(.......)))).((((....)))).)))))---)).(((((..((((.....((((.....))))...))))..)))))..... ( -29.20, z-score = -1.21, R) >droSec1.super_1 14003652 104 - 14215200 ---CAGCACCCCCACCCACCAAUUU---------CCCCCCAAGAAGAUGCGAACGCGCAAGGUGC---UGUUGGUAAUUGGCUUUCUGGUUACGUGGACCUAUGCCACCUACUACCUGC ---(((((((............(((---------(.......)))).((((....)))).)))))---)).(((((..((((.....((((.....))))...))))..)))))..... ( -29.20, z-score = -1.31, R) >droEre2.scaffold_4845 10593374 107 - 22589142 CUAAGCCACAUACACCCACCAAUUC---------CCCCCCAAGAAGAUGCGAACGCGCAAGGUGC---UGUUGGUAAUUGGGUUUCUGGUUACGUGGACCUAUGCCACCUACUACCUGC ....((((.(((((((......(((---------........)))..((((....)))).)))).---)))))))....((((....(((..((((....))))..)))....)))).. ( -23.10, z-score = 1.23, R) >droAna3.scaffold_13266 15728532 102 + 19884421 ------------CGUAGUCUUAAUCGAAAUCGAAGCCG--AACAAAAUGCGGUCGCGCAAGGUGC---UUUUGGUGAUCGGUUUUCUGGUUACGUGGACAUACGCCACCUACUACCUUC ------------.(((((..(((((((((((((.((((--((((...((((....))))...)).---.))))))..))))))))..))))).((((.(....)))))..))))).... ( -28.20, z-score = -0.77, R) >dp4.chr3 6308675 86 + 19779522 -----------------ACCAAACC---------CCCC----AAAGAUGCGAUCGCGUAAGGUGC---UGCUUAUAGUUGGUUUUCUGAUUACGUGGACCUACGCCACCUACUACCUAC -----------------........---------....----............(((((.(((.(---..(...((((..(....)..)))).)..))))))))).............. ( -16.50, z-score = -0.08, R) >droPer1.super_2 8149525 86 - 9036312 -----------------ACCAAACC---------CCCC----AAAGAUGCGAUCGCGUAAGGUGC---UGCUUAUAGUUGGUUUUCUGAUUACGUGGACCUACGCCACCUACUACCUAC -----------------........---------....----............(((((.(((.(---..(...((((..(....)..)))).)..))))))))).............. ( -16.50, z-score = -0.08, R) >droWil1.scaffold_180700 3135303 113 + 6630534 ---CACCACCCCCGCCCACCCCUUUUGGAACAAGCCCAGUUCGAAGAUGCGUUCCCGCAAAGUGC---UGUUAGUGAUUGGUUUUUUGAUUACGUGGACCUAUGUCACCUAUUACAUAU ---..((((....((.(.....((((.((((.......)))).))))((((....))))..).))---.....(((((..(....)..)))))))))...(((((........))))). ( -23.60, z-score = -0.82, R) >droVir3.scaffold_12875 13641196 84 - 20611582 ----------------------------------CACA-CAGCAAUAUGCGUUCGCGCAAGGUGCAGGUGAUGGUGAUUAGCUUCCUCAUCAUGUGGACGUUCAUUACGUAUUAUAUAC ----------------------------------..((-(.(((...((((....))))...)))..(((((((.((......)).)))))))))).((((.....))))......... ( -23.60, z-score = -1.19, R) >droMoj3.scaffold_6496 9063050 104 - 26866924 --------------CACACCCACACAUAAACACAGACG-UUGCAAAAUGCGUACACGUAAGGUACAAAUUUUGGUUAUUGGCGUCCUCAUCCUAUGGACAUUCGUCACGUACUACAUAU --------------....................((((-...((((((..((((.(....)))))..)))))).........((((.........))))...))))............. ( -20.40, z-score = -0.41, R) >consensus ____________C_CCCACCAAUUC_________CCCC_CAAGAAGAUGCGAACGCGCAAGGUGC___UGUUGGUAAUUGGCUUUCUGAUUACGUGGACCUACGCCACCUACUACCUAC ...............................................((((....)))).((((.........((((((((....))))))))((((.......)))).))))...... (-12.46 = -11.27 + -1.19)

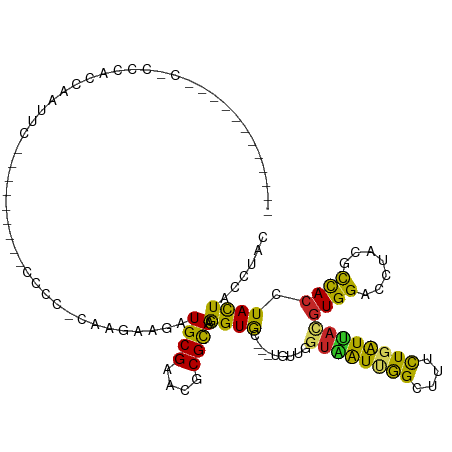
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:36 2011