| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,414,208 – 16,414,272 |
| Length | 64 |
| Max. P | 0.500000 |

| Location | 16,414,208 – 16,414,272 |
|---|---|
| Length | 64 |
| Sequences | 9 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 70.91 |
| Shannon entropy | 0.58897 |
| G+C content | 0.57292 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -9.45 |
| Energy contribution | -8.99 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.82 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
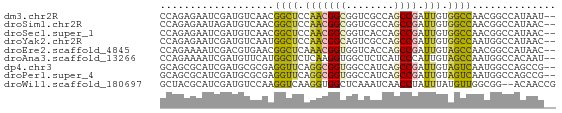
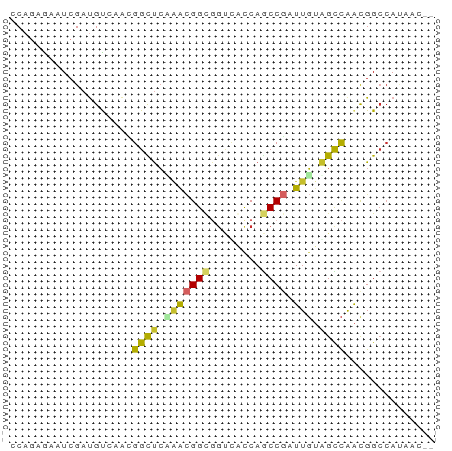
>dm3.chr2R 16414208 64 + 21146708 CCAGAGAAUCGAUGUCAACGGCUCCAACGGCGGUCGCCAGCCGAUUGUGGCCAACGGCCAUAAU-- ..................(((((.....(((....))))))))(((((((((...)))))))))-- ( -22.10, z-score = -1.38, R) >droSim1.chr2R 15073297 64 + 19596830 CCAGAGAAUAGAUGUCAACGGCUCCAACGGCGGUCGCCAGCCGAUUGUGGCCAACGGCCAUAAC-- ..................(((((.....(((....)))))))).((((((((...)))))))).-- ( -21.40, z-score = -1.57, R) >droSec1.super_1 13969860 64 + 14215200 CCAGAGAAUCGAUGUCAACGGCUCCAACGGCGGUCACCAGCCGAUUGUGGCCAACGGCCAUAAC-- ...(.((..((.......))..)))..((((........)))).((((((((...)))))))).-- ( -19.50, z-score = -1.01, R) >droYak2.chr2R 8356631 64 - 21139217 CCAGAGAAUCGAUGUCAAUGGCUCCAACGGCAGUCGCCAGCCGAUUGUGGCCAAUGGCCAUAAC-- .............((((.((((..(((((((........)))).)))..)))).))))......-- ( -22.00, z-score = -2.07, R) >droEre2.scaffold_4845 10559287 64 + 22589142 CCAGAAAAUCGACGUGAACGGCUCAAACGGUGGUCACCAGCCGAUUGUAGCCAACGGCCAUAAC-- ............(((....(((((((.((((((...))).))).))).)))).)))........-- ( -16.60, z-score = -0.90, R) >droAna3.scaffold_13266 17017908 64 - 19884421 CCAGAAAAUCGAUGUUCAUGGCUCUCAAGGUGGCUCUCAUCCCAUUGUAGCCAAUGGCCACAAU-- ..........(.((..(((((((.....(((((........)))))..)))).)))..)))...-- ( -12.70, z-score = 0.28, R) >dp4.chr3 10500520 64 + 19779522 GCAGCGCAUCGAUGCGCGAGGUUCAGGCGGUGGCCAUCAGCCGAUUGUAGUCAAUGGCCAGCCG-- ...(((((....)))))........(((..(((((((..((........))..)))))))))).-- ( -24.50, z-score = -0.98, R) >droPer1.super_4 5828298 64 + 7162766 GCAGCGCAUCGAUGCGCGAGGUUCAGGCGGUGGCCAUCAGCCGAUUGUAGUCAAUGGCCAGCCG-- ...(((((....)))))........(((..(((((((..((........))..)))))))))).-- ( -24.50, z-score = -0.98, R) >droWil1.scaffold_180697 294404 64 - 4168966 GCUACGCAUCGAUGUCCAAGGUCAAGGUGGCUCAAAUCAACCUAUUUAUGUUGGCGG--ACAACCG ............(((((..((((.....))))....(((((........))))).))--))).... ( -12.40, z-score = 0.38, R) >consensus CCAGAGAAUCGAUGUCAACGGCUCAAACGGCGGUCACCAGCCGAUUGUAGCCAACGGCCAUAAC__ ...................((((.(((((((........)))).))).)))).............. ( -9.45 = -8.99 + -0.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:33 2011