
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,388,199 – 16,388,312 |
| Length | 113 |
| Max. P | 0.520543 |

| Location | 16,388,199 – 16,388,312 |
|---|---|
| Length | 113 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 75.48 |
| Shannon entropy | 0.48749 |
| G+C content | 0.39046 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -11.68 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.520543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16388199 113 - 21146708 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG--GUUGACUAUCUAGCUGGCUUGAGGUAAGCCUUUUGUU ((((((((.....(((((.....))))).............------...........((....))....))))))))((((--(........))))).((((((...))))))....... ( -29.70, z-score = -1.69, R) >droPer1.super_4 5800688 111 - 7162766 GAGGCGUAAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUACUAAAUAAAAAUAGAAUAGAUAAAUCCGCCCACGAAUGGGUGG-AUUCCCCCUCUGUUUGGGCAUCCUUAUU--------- ((((.((......(((((.....))))).........................(((((((..((((((((((....)))))))-))).....)))))))..))..))))...--------- ( -28.90, z-score = -2.50, R) >dp4.chr3 10472898 111 - 19779522 GAGGCGUAAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUACUAAAUAAAAAUAGAAUAGAUAAAUCCGCCCACGAAUGGGGUG-AUUCCCCCUCUGUUUGGGCAUCCUUAUU--------- ((((.........(((((.....))))).......................................(((((.....((((..-....))))......)))))..))))...--------- ( -25.70, z-score = -1.49, R) >droAna3.scaffold_13266 16990479 109 + 19884421 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUGGAUAAAUCCGCCCACGGCUCGCUG--GUUGGCUAUC----UGGCUUGAGGUAAGCCUUUUGUU .(((((((.....(((((.....))))).............------..........(((....)))...))))))).((..--(........)----..))..((((....))))..... ( -26.90, z-score = -0.72, R) >droEre2.scaffold_4845 10533238 113 - 22589142 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG--GUUGACUAUCUAGCUGGCUUGAGGUAAGCCUUUUGUU ((((((((.....(((((.....))))).............------...........((....))....))))))))((((--(........))))).((((((...))))))....... ( -29.70, z-score = -1.69, R) >droYak2.chr2R 8330023 113 + 21139217 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG--GUUGACUAUCUAGCUGGCUUGAGGUAAGCCUUUUGUU ((((((((.....(((((.....))))).............------...........((....))....))))))))((((--(........))))).((((((...))))))....... ( -29.70, z-score = -1.69, R) >droSec1.super_1 13944141 113 - 14215200 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG--GUUGACUAUCUAGGUGGCUUGAGGUAAGCCUUUUGUU ((((((((.....(((((.....))))).............------...........((....))....)))))))).(((--(........))))..((((((...))))))....... ( -26.60, z-score = -0.64, R) >droSim1.chr2R 15045843 113 - 19596830 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG--GUUGACUAUCUAGCUGGCUUGAGGUAAGCCUUUUGUU ((((((((.....(((((.....))))).............------...........((....))....))))))))((((--(........))))).((((((...))))))....... ( -29.70, z-score = -1.69, R) >droWil1.scaffold_180697 246235 89 + 4168966 CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC------AAAAUAGAAGAGAUAAAUCCCCCUA--GCCGGUAG--GUAAGCCCUUUUGUU---------------------- .....((((....)))).....((((((.........))))------))((((((((.(....(((..((..--...))..)--))...).))))))))---------------------- ( -12.30, z-score = -0.35, R) >droVir3.scaffold_13284 2017 93 + 3046 CAGCCGUGAAUAAUCGCAUUAGUUACGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCGCCCACGGUUGGGCGGAGGUAAGCCUUUUGUU---------------------- ....(((((.((((...)))).)))))............((------((((.............((((((((....))))))))((....)))))))).---------------------- ( -22.80, z-score = -2.00, R) >droMoj3.scaffold_6496 18411245 93 - 26866924 CAGCCGUGAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCACCCACGGCUGGGCGCCGCUAAGCCUUUUGUU---------------------- ((((((((.....(((((.....))))).............------...........((....))....))))))))(((........))).......---------------------- ( -24.70, z-score = -2.50, R) >droGri2.scaffold_15245 16155955 93 + 18325388 CAGCUGUGAAUAAUCGCAUUAGUUGCGAAAUUAAAAAUUAC------AAAAUAGAAUAGAUAAAUCCACCCACGGCUGGUCGGAGGUAAGCUCUUUGUU---------------------- ((((((((.....(((((.....))))).............------...........((....))....))))))))...((((.....)))).....---------------------- ( -20.30, z-score = -1.45, R) >consensus CAGCCGUGAAUAAUCGCAUUAGUUGUGAAAUUAAAAAUUAC______AAAAUAGAAUAGAUAAAUCCGCCCACGGCUGGCUG__GUUGACCAUCUAGUUGGCUUGAGGUAAG_________ .(((((((.....(((((.....)))))..............................((....))....)))))))............................................ (-11.68 = -12.08 + 0.40)

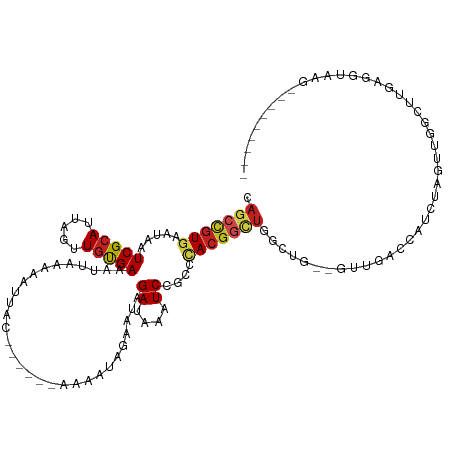
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:32 2011