| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,376,504 – 16,376,614 |
| Length | 110 |
| Max. P | 0.747263 |

| Location | 16,376,504 – 16,376,614 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Shannon entropy | 0.32531 |
| G+C content | 0.48168 |
| Mean single sequence MFE | -34.96 |
| Consensus MFE | -19.32 |
| Energy contribution | -20.01 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739352 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
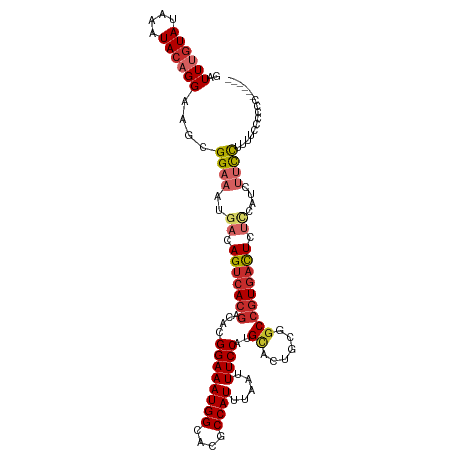
>dm3.chr2R 16376504 110 + 21146708 ---GGGGGGGAAGAAAGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC ---..(.((((((...(((((...((.(((((((((.((...))((((((.....((((....))))))))))))))))))).)).))))).)))))).)............. ( -40.50, z-score = -3.16, R) >droSim1.chr2R 15033596 108 + 19596830 -----GGGGGGGAAAAGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC -----.((((((....(((((...((.(((((((((.((...))((((((.....((((....))))))))))))))))))).)).))))).))))))............... ( -38.20, z-score = -2.57, R) >droSec1.super_1 13932782 108 + 14215200 -----GGGGGGGAAAAGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC -----.((((((....(((((...((.(((((((((.((...))((((((.....((((....))))))))))))))))))).)).))))).))))))............... ( -38.20, z-score = -2.57, R) >droYak2.chr2R 8318176 107 - 21139217 ------GGGGGGAAAAGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC ------.(.(((((..(((((...((.(((((((((.((...))((((((.....((((....))))))))))))))))))).)).)))))..))))).)............. ( -38.00, z-score = -2.77, R) >droEre2.scaffold_4845 10521622 107 + 22589142 ------GGGGGAAAGCGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC ------..(((((.(((((((...((.(((((((((.((...))((((((.....((((....))))))))))))))))))).)).))))))))))))............... ( -42.60, z-score = -3.94, R) >droAna3.scaffold_13266 16979019 107 - 19884421 ------GAGGCAGACAGAGAUAGGAAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGUUUCCUGUAUUUAUACAAAUC ------.(((.((((.(((((.((...(((((((((.((...))((((((.....((((....))))))))))))))))))).))))))).)))))))............... ( -32.00, z-score = -1.89, R) >dp4.chr3 10448929 113 + 19779522 GAGAGAGAGACCCAGCAGACCCGUCGCAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGCCGUGACUGUUAUUUCCGUUUCCGGUAUUUAUACAAAUC ....(.(((((......((....))(((((((((((.((...))((((((.....((((....))))))))))))))))))))).......))))))................ ( -33.30, z-score = -1.72, R) >droPer1.super_4 5783461 113 + 7162766 GAGAGAGAGACCCAGCAGACGCGUCGCAGUCACGGCCGCAGUACAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGCAGUGACUGUUAUUUCCGUUUCCGGUAUUUAUAAAAAUC ....(.(((((...((....))...((((((((.((....)).(((((((.....((((....)))))))))))..)))))))).......))))))................ ( -27.60, z-score = -0.16, R) >droGri2.scaffold_15245 16140756 99 - 18325388 ------------GAAAGAGACAGAGAGAA--AUGGAAUGGAUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC ------------...(((.((((..((..--..(((((((...(((((.....((((((....)))))).....)))))....)))))))..))..)))).)))......... ( -24.20, z-score = -1.07, R) >consensus ______GGGGGAAAAAGGAAGAUGGAGAGUCACGGCCGCAGUGCAUGGAAAUUAAAUGGCGUGCCAUUUCCGUGUCGUGACUGUCAUUUCCGCUUCCUGUAUUUAUACAAAUC ................((((.....(.(((((((........((((((((.....((((....))))))))))))))))))).)...))))...................... (-19.32 = -20.01 + 0.69)



| Location | 16,376,504 – 16,376,614 |
|---|---|
| Length | 110 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Shannon entropy | 0.32531 |
| G+C content | 0.48168 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -17.48 |
| Energy contribution | -18.66 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.747263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16376504 110 - 21146708 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCUUUCUUCCCCCCC--- ....((((....))))(((((.(((((((..(((((((....((((((((....)))).....))))..((......)))))))))...))).))))...))))).....--- ( -29.00, z-score = -2.15, R) >droSim1.chr2R 15033596 108 - 19596830 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCUUUUCCCCCCC----- ....((((....))))(((((.(((((((..(((((((....((((((((....)))).....))))..((......)))))))))...))).)))).))))).....----- ( -29.10, z-score = -2.26, R) >droSec1.super_1 13932782 108 - 14215200 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCUUUUCCCCCCC----- ....((((....))))(((((.(((((((..(((((((....((((((((....)))).....))))..((......)))))))))...))).)))).))))).....----- ( -29.10, z-score = -2.26, R) >droYak2.chr2R 8318176 107 + 21139217 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCUUUUCCCCCC------ ....((((....))))(((((.(((((((..(((((((....((((((((....)))).....))))..((......)))))))))...))).)))).)))))....------ ( -29.10, z-score = -2.29, R) >droEre2.scaffold_4845 10521622 107 - 22589142 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCGCUUUCCCCC------ ....((((....))))(((((((((((((..(((((((....((((((((....)))).....))))..((......)))))))))...))).))))))))))....------ ( -34.60, z-score = -3.72, R) >droAna3.scaffold_13266 16979019 107 + 19884421 GAUUUGUAUAAAUACAGGAAACGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUUCCUAUCUCUGUCUGCCUC------ ....((((....))))((..(((((.(((..(((((((....((((((((....)))).....))))..((......))))))))).....))).)))))...))..------ ( -24.30, z-score = -0.72, R) >dp4.chr3 10448929 113 - 19779522 GAUUUGUAUAAAUACCGGAAACGGAAAUAACAGUCACGGCACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUGCGACGGGUCUGCUGGGUCUCUCUCUC (((((((.......(((....)))......((((((((((..((((((((....)))).....)))).(((...)))))))))))))..)))))))....(((.....))).. ( -40.20, z-score = -3.37, R) >droPer1.super_4 5783461 113 - 7162766 GAUUUUUAUAAAUACCGGAAACGGAAAUAACAGUCACUGCACGGAAAUGGCACGCCAUUUAAUUUCCAUGUACUGCGGCCGUGACUGCGACGCGUCUGCUGGGUCUCUCUCUC ..............(((....)))......(((((((.((..((((((((....)))).....)))).((.....)))).))))))).(((.((.....)).)))........ ( -30.60, z-score = -1.35, R) >droGri2.scaffold_15245 16140756 99 + 18325388 GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCAUCCAUUCCAU--UUCUCUCUGUCUCUUUC------------ (((.((((....))))(((.((((((((....((....))....((((((....)))))).))))))..)).))).......--........)))......------------ ( -18.00, z-score = -0.20, R) >consensus GAUUUGUAUAAAUACAGGAAGCGGAAAUGACAGUCACGACACGGAAAUGGCACGCCAUUUAAUUUCCAUGCACUGCGGCCGUGACUCUCCAUCUUCCUUUUCCCCCC______ ..((((((....))))))....((((..((.(((((((....((((((((....)))).....))))..((......))))))))).))....))))................ (-17.48 = -18.66 + 1.18)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:31 2011