| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,375,207 – 16,375,306 |
| Length | 99 |
| Max. P | 0.997208 |

| Location | 16,375,207 – 16,375,306 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 67.15 |
| Shannon entropy | 0.65045 |
| G+C content | 0.45538 |
| Mean single sequence MFE | -23.76 |
| Consensus MFE | -13.36 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.997208 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
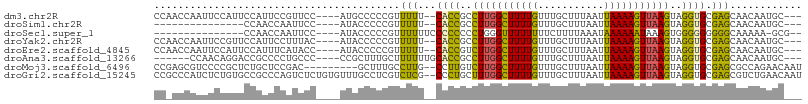
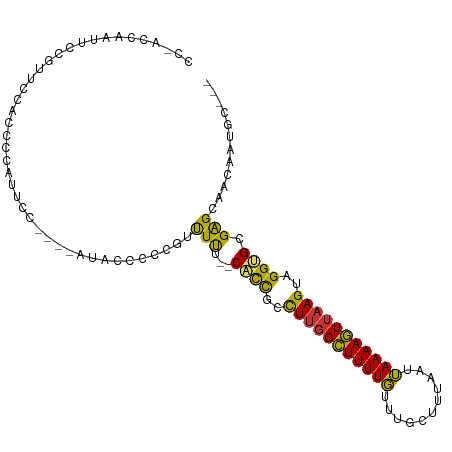
>dm3.chr2R 16375207 99 - 21146708 CCAACCAAUUCCAUUCCAUUCCGUUCC----AUGCCCCCGUUUUU--CACCGCCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC--- ...........................----........((((..--((((..(((((((((((...........)))))))))))..)))).))))........--- ( -18.10, z-score = -1.21, R) >droSim1.chr2R 15032319 84 - 19596830 ---------------CCAACCAAUUCC----AUACCCCCGUUUUU--CACCGCCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC--- ---------------............----........((((..--((((..(((((((((((...........)))))))))))..)))).))))........--- ( -18.10, z-score = -1.72, R) >droSec1.super_1 13931504 86 - 14215200 ---------------CCAACCAAUUCC----AUACCCCCGUUUUUUCCCCCCCCUGGGUUUUUUUUCUUUUAAAUAAAAAAUAAAGUGGGGGGGGGCAAAAA-GCG-- ---------------............----.......((((((((((((((((((.(((((((...........)))))))....)))))))))).)))))-)))-- ( -29.40, z-score = -2.16, R) >droYak2.chr2R 8316839 99 + 21139217 CCAACCAAUUCCGUUCCAUUCCUUUAC----AUACCCCCGUUUUU--CACCGCCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC--- ...........................----........((((..--((((..(((((((((((...........)))))))))))..)))).))))........--- ( -18.10, z-score = -1.53, R) >droEre2.scaffold_4845 10520312 99 - 22589142 CCAACCAAUUCCAUUCCAUUUCAUACC----AUACCCCCGUUUUU--CACCGUCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC--- ...........................----........((((..--((((..(((((((((((...........)))))))))))..)))).))))........--- ( -17.40, z-score = -1.66, R) >droAna3.scaffold_13266 16977789 95 + 19884421 ------CCAACAGGACCGCCCCUGCCC----CCGCUUUGCUUUUUUGCACCGCCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC--- ------....((((......))))...----..((.(((.((.((((((((..(((((((((((...........)))))))))))..)))))))).))))).))--- ( -29.10, z-score = -2.22, R) >droMoj3.scaffold_6496 18389792 97 - 26866924 CCGAGCGUCCCCGCUCUGCUCCGAC---------GCUUUGCCUUG--CCUUGUCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCGCCAGAACAAU ..(((((((...((...))...)))---------)))).((((((--(((((.(((((((((((...........))))))))))))))).))))).))......... ( -30.50, z-score = -2.15, R) >droGri2.scaffold_15245 16139106 106 + 18325388 CCGCCCAUCUCUGUGCCGCCCAGUCUCUGUGUUUGCCUCGUCUCG--CCCUGCUUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCGUCUGAACAAU ..((.((....)).)).............((((((...((.((((--((((((((.((((((((...........))))))))))))))).)))))))..)))))).. ( -29.40, z-score = -2.74, R) >consensus CC_ACCAAUUCCGUUCCACCCCAUUCC____AUACCCCCGUUUUU__CACCGCCUUGGCUUUUGUUUGCUUUAAUUAAAAGUUAAGUAGGUGCGAGCAACAAUGC___ .......................................((((....((((..(((((((((((...........)))))))))))..)))).))))........... (-13.36 = -13.05 + -0.31)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:29 2011