
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,327,762 – 16,327,814 |
| Length | 52 |
| Max. P | 0.772617 |

| Location | 16,327,762 – 16,327,814 |
|---|---|
| Length | 52 |
| Sequences | 5 |
| Columns | 69 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.36955 |
| G+C content | 0.39853 |
| Mean single sequence MFE | -13.75 |
| Consensus MFE | -7.94 |
| Energy contribution | -8.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772617 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
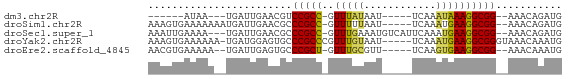
>dm3.chr2R 16327762 52 + 21146708 ------AUAA---UGAUUGAACGUCCGCC-GUUUAUAAU-----UCAAAUAAAGGCGG--AAACAGAUG ------....---..(((.....((((((-.(((((...-----....))))))))))--)....))). ( -12.80, z-score = -2.77, R) >droSim1.chr2R 14984114 61 + 19596830 AAAGUGAAAAAAAUGAUUGAACGCCCGCC-GUUUUUAAU-----UCAAAUGAAGGCGG--AAACAGAUG ........................(((((-.........-----.........)))))--......... ( -8.87, z-score = -0.25, R) >droSec1.super_1 13884058 63 + 14215200 AAAUUGAAAA---UGAUUGAACGCCCGCC-GUUUGAAAUGUCAUUCAAAUGAAGGCGG--AAACAGAUG ..........---...........(((((-(((((((......)))))))...)))))--......... ( -13.60, z-score = -1.56, R) >droYak2.chr2R 8266757 63 - 21139217 AAAGUGAAAAAA-UGAUGGAGUGCCCGCCCGUUUGUAAU-----UCAAAUGAAGGCGGGUAAACAAAUG ............-...((...((((((((((((((....-----.))))))..))))))))..)).... ( -20.90, z-score = -4.37, R) >droEre2.scaffold_4845 10473426 59 + 22589142 AACGUGAAAAA--UGAUUGAGUGCCCGCU-GUUUGCGUU-----UCAAGUGAAGGCGG--AAACAAAUG ..(((.....)--)).....((..(((((-.((..(...-----....)..)))))))--..))..... ( -12.60, z-score = -0.67, R) >consensus AAAGUGAAAAA__UGAUUGAACGCCCGCC_GUUUGUAAU_____UCAAAUGAAGGCGG__AAACAGAUG ........................(((((.(((((..........)))))...)))))........... ( -7.94 = -8.02 + 0.08)

| Location | 16,327,762 – 16,327,814 |
|---|---|
| Length | 52 |
| Sequences | 5 |
| Columns | 69 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Shannon entropy | 0.36955 |
| G+C content | 0.39853 |
| Mean single sequence MFE | -11.30 |
| Consensus MFE | -6.32 |
| Energy contribution | -6.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638293 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
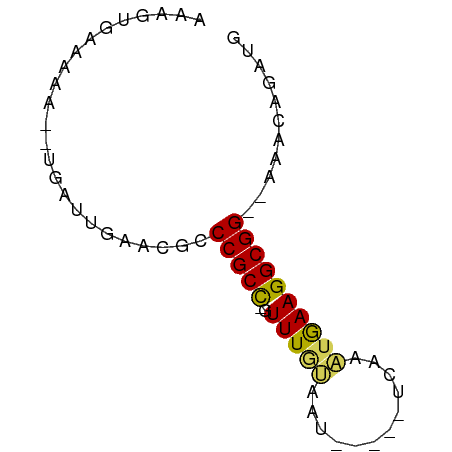
>dm3.chr2R 16327762 52 - 21146708 CAUCUGUUU--CCGCCUUUAUUUGA-----AUUAUAAAC-GGCGGACGUUCAAUCA---UUAU------ ........(--((((((((((....-----...))))).-))))))..........---....------ ( -12.40, z-score = -2.90, R) >droSim1.chr2R 14984114 61 - 19596830 CAUCUGUUU--CCGCCUUCAUUUGA-----AUUAAAAAC-GGCGGGCGUUCAAUCAUUUUUUUCACUUU ........(--((((((((....))-----)........-))))))....................... ( -8.40, z-score = -0.66, R) >droSec1.super_1 13884058 63 - 14215200 CAUCUGUUU--CCGCCUUCAUUUGAAUGACAUUUCAAAC-GGCGGGCGUUCAAUCA---UUUUCAAUUU ........(--(((((....((((((......)))))).-))))))..........---.......... ( -13.00, z-score = -1.87, R) >droYak2.chr2R 8266757 63 + 21139217 CAUUUGUUUACCCGCCUUCAUUUGA-----AUUACAAACGGGCGGGCACUCCAUCA-UUUUUUCACUUU ..........(((((((...((((.-----....)))).)))))))..........-............ ( -14.00, z-score = -2.95, R) >droEre2.scaffold_4845 10473426 59 - 22589142 CAUUUGUUU--CCGCCUUCACUUGA-----AACGCAAAC-AGCGGGCACUCAAUCA--UUUUUCACGUU .........--..((((((....))-----).(((....-.)))))).........--........... ( -8.70, z-score = -0.75, R) >consensus CAUCUGUUU__CCGCCUUCAUUUGA_____AUUACAAAC_GGCGGGCGUUCAAUCA__UUUUUCACUUU ...........(((((........................)))))........................ ( -6.32 = -6.52 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:25 2011