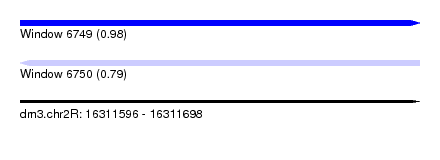
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,311,596 – 16,311,698 |
| Length | 102 |
| Max. P | 0.980888 |

| Location | 16,311,596 – 16,311,698 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 74.76 |
| Shannon entropy | 0.48181 |
| G+C content | 0.58953 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.75 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.980888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
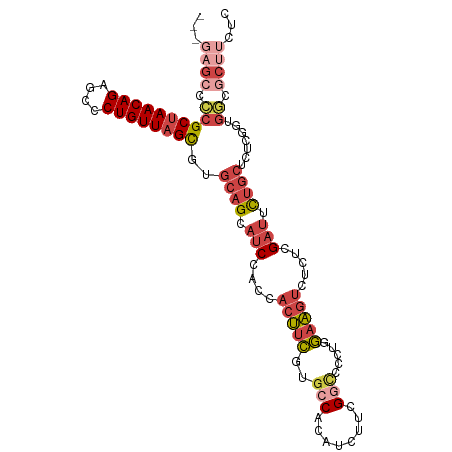
>dm3.chr2R 16311596 102 + 21146708 ---GAGCACCGCUAACAGAGCCCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAACUUCGGCCACUUGAAGUCUCUCGAUUUUGCUCUCGGUGGCGCUUCUC ---..((((.((((((((....))))))))))))(((..(((((.....(((((........)).)))..((((((....))))))......))))).))).... ( -37.10, z-score = -3.31, R) >droSim1.chr2R 14967987 102 + 19596830 ---GAGCCCCGCUAACAGAGCCCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAUCUUCGGCCGCUGGAAGUCUCUCGAUUCUGCUCUCGGUGGCGCUUCUC ---..(((((((((((((....))))))))(.((((.(((....((((((((((........)).)))..))))).....))).)))).)..)).)))....... ( -36.50, z-score = -2.08, R) >droSec1.super_1 13867949 102 + 14215200 ---GAGCCCCGCUAACAGAGCCCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAUCUUCGUCCGCUGGAAGUCUCUCGAUUCUGCUCUCGGUGGCGCUUCUC ---..(((((((((((((....))))))))(.((((.(((....(((((..((.((......))..))..))))).....))).)))).)..)).)))....... ( -33.80, z-score = -1.99, R) >droYak2.chr2R 8250014 102 - 21139217 ---GCGAGGCGCUAACAGCGCGCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAUCUUCGGCACGUUGAAGUCUCUCGAUUUUGCUCUCGUUGCCGCUUCUC ---..(((((((((((((....))))))))(.(((((...........((((((........))))))..((((((....))))))......))))))))))).. ( -40.60, z-score = -3.45, R) >droEre2.scaffold_4845 10456903 102 + 22589142 ---GAGCCGCGCUAACAGAGCGCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAUCUUCGGUCCGUUGAAGUCUCUCGAUGUUGCUCUCGGUGCCGCUGCUC ---((((.((((((((((....))))))))(.((((((((....((((((.(((........)))....)))))).....)))))))).)........)).)))) ( -39.20, z-score = -2.25, R) >dp4.chr3 10376455 87 + 19779522 GGGUACCACUGCUAACAGAGCUCUGUU-GUCCACAGUCUCCGUUUCGUGAUGCCAGCGCAUCGUGCCAUGCCCUCGUCACGUCUCGUC----------------- (((((.(((.((.(((.(((..((((.-....)))).))).)))..))(((((....))))))))...)))))...............----------------- ( -22.30, z-score = -0.17, R) >consensus ___GAGCCCCGCUAACAGAGCCCUGUUAGCGUGCAGCAUCCACCACUUCGUGCCACAUCUUCGGCCCCUGGAAGUCUCUCGAUUCUGCUCUCGGUGGCGCUUCUC .....((.((((((((((....))))))))(.((((.(((....(((((..(((........))).....))))).....))).)))).).....)).))..... (-19.14 = -20.75 + 1.61)

| Location | 16,311,596 – 16,311,698 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 74.76 |
| Shannon entropy | 0.48181 |
| G+C content | 0.58953 |
| Mean single sequence MFE | -37.55 |
| Consensus MFE | -18.91 |
| Energy contribution | -21.05 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.788729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16311596 102 - 21146708 GAGAAGCGCCACCGAGAGCAAAAUCGAGAGACUUCAAGUGGCCGAAGUUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGGGCUCUGUUAGCGGUGCUC--- ....(((((((((..((......)).....(((((..(((.(((......)))))))))))))))).))))((((((((((((....)))))))).))))..--- ( -40.20, z-score = -2.27, R) >droSim1.chr2R 14967987 102 - 19596830 GAGAAGCGCCACCGAGAGCAGAAUCGAGAGACUUCCAGCGGCCGAAGAUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGGGCUCUGUUAGCGGGGCUC--- ....((((((((((...((.(((((....)).)))..)).((((......))))......)))))).))))((.(((((((((....)))))))))..))..--- ( -40.10, z-score = -1.68, R) >droSec1.super_1 13867949 102 - 14215200 GAGAAGCGCCACCGAGAGCAGAAUCGAGAGACUUCCAGCGGACGAAGAUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGGGCUCUGUUAGCGGGGCUC--- ....((((((((((.(.((....((....))((((........)))).....)).)....)))))).))))((.(((((((((....)))))))))..))..--- ( -36.60, z-score = -1.17, R) >droYak2.chr2R 8250014 102 + 21139217 GAGAAGCGGCAACGAGAGCAAAAUCGAGAGACUUCAACGUGCCGAAGAUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGCGCGCUGUUAGCGCCUCGC--- (((..((((((.(.(..((....((....))......(((((((......)))))))..))..).).)))))).(((((((((....))))))))).)))..--- ( -42.80, z-score = -2.86, R) >droEre2.scaffold_4845 10456903 102 - 22589142 GAGCAGCGGCACCGAGAGCAACAUCGAGAGACUUCAACGGACCGAAGAUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGCGCUCUGUUAGCGCGGCUC--- ((((.(((((((((...((....((....))......((..(((......)))..))..))..))).)))))).(((((((((....)))))))))..))))--- ( -38.70, z-score = -1.43, R) >dp4.chr3 10376455 87 - 19779522 -----------------GACGAGACGUGACGAGGGCAUGGCACGAUGCGCUGGCAUCACGAAACGGAGACUGUGGAC-AACAGAGCUCUGUUAGCAGUGGUACCC -----------------...............(((.((.((..(((((....)))))....(((((((.((.((...-..)).)))))))))....)).)).))) ( -26.90, z-score = -0.71, R) >consensus GAGAAGCGCCACCGAGAGCAAAAUCGAGAGACUUCAAGGGGCCGAAGAUGUGGCACGAAGUGGUGGAUGCUGCACGCUAACAGGGCUCUGUUAGCGGGGCUC___ .....((.((.......(((..(((.(...(((((.....((((......))))..)))))..).)))..)))..((((((((....)))))))))).))..... (-18.91 = -21.05 + 2.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:21 2011