
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,287,979 – 16,288,071 |
| Length | 92 |
| Max. P | 0.809153 |

| Location | 16,287,979 – 16,288,071 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 78.31 |
| Shannon entropy | 0.46646 |
| G+C content | 0.44151 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -16.28 |
| Energy contribution | -16.39 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16287979 92 + 21146708 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGUUGCGGGGAC---GGCUGCUAGCCUGCU------------ (((((((((((.....))))))))))).............((((((-..((((((..((.....))..))))))...)---)))))..........------------ ( -26.70, z-score = -1.41, R) >droSim1.chr2R 14942813 92 + 19596830 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGUUGCGGGGAC---GGCUGCUAGCCUGCU------------ (((((((((((.....))))))))))).............((((((-..((((((..((.....))..))))))...)---)))))..........------------ ( -26.70, z-score = -1.41, R) >droSec1.super_1 13844674 92 + 14215200 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGUUGCGGGGAC---GGCUGCUAGCCCGCU------------ (((((((((((.....))))))))))).............((((((-..((((((..((.....))..))))))...)---)))))..........------------ ( -26.70, z-score = -1.32, R) >droYak2.chr2R 8225702 92 - 21139217 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGUUGCGGGGAG---UUCUGCUAGCUGGCU------------ (((((((((((.....))))))))))).............((((((-..((((((..((.....))..))))))((..---..))..))))))...------------ ( -26.00, z-score = -1.19, R) >droEre2.scaffold_4845 10431999 92 + 22589142 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGCUGUGGGGAG---GGCCGCUAGCCUGCU------------ (((((((((((.....)))))))))))...........((((((..-.....((....)).......)))))).((.(---(((.....)))).))------------ ( -27.14, z-score = -1.03, R) >droAna3.scaffold_13266 16880752 102 - 19884421 UGUCUGUUUGUUAUAGCCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGCAAACUUGAUUAAAUGUUGCCGCAGCG-AGAUGGCGAAACGGUGAGACGACC---- ((((((((((.......))))))))))...........(((.((((-.(((((.(((..........)))...))))).-...))))......)))........---- ( -23.20, z-score = -0.46, R) >dp4.chr3 10349189 84 + 19779522 UCUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCGG-AGCUGCAAACUUGAUUAAAUGCUGCCGCAGCG-CGCUGG---------------------- ..(((((((((.....)))))))))...............(((((.-.(((((....................))))).-))))).---------------------- ( -24.65, z-score = -1.57, R) >droPer1.super_4 5683067 84 + 7162766 UCUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCGG-AGCUGCAAACUUGAUUAAAUGCUGCCGCAGCG-CGCUGG---------------------- ..(((((((((.....)))))))))...............(((((.-.(((((....................))))).-))))).---------------------- ( -24.65, z-score = -1.57, R) >droWil1.scaffold_180697 4096004 104 + 4168966 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG-AGCUGAAAACUUGAUUAAAUGCUACCGCAUCU-CAG-AGCGAAACGGAUAUAAAGUUAAA- (((((((((((.....))))))))))).(((((.......((((..-.))))...................(((..((.-...-...))..))).......))))).- ( -22.50, z-score = -1.52, R) >droVir3.scaffold_12875 11674305 97 - 20611582 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCCA-AGCUGAAAACUUGAUUAAAUG-UGCCGCAGCGACAGCAGCAGUCAGGGUCCU--------- (((((((((((.....)))))))))))((((((.....((((..((-((.(....)))))......))-))..((.((....)).)).)))))).....--------- ( -29.60, z-score = -2.18, R) >droMoj3.scaffold_6496 18242159 107 + 26866924 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACAUAGCCGGAGCUGAAAACUUGAUUAAAUG-UGCCGCAGCAGCGACAGCAGCAACAGCAGUCACGAUGCU (((((((((((.....)))))))))))(.((((.........((.(..((((...((..........)-).......))))..).)).((....)).)))).)..... ( -27.20, z-score = -0.24, R) >droGri2.scaffold_15245 16031053 97 - 18325388 UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACAUAGCUG-AGCUGAAAACUUGAUUAAAUG-UGCCGCAGCGACAGCAGCAGUCAGGGUCCU--------- (((((((((((.....)))))))))))((((((.((....))((((-.((((...((..........)-)....))))..))))....)))))).....--------- ( -30.10, z-score = -2.17, R) >consensus UGUCUGUUUGUUAUAGGCAAAUAGACACUUGACAUAAACACAGCUG_AGCUGCAAACUUGAUUAAAUGUUGCCGCAGC__CGGCAGCUAGCCGGCU____________ (((((((((((.....))))))))))).((((((......((((....))))......)).))))........................................... (-16.28 = -16.39 + 0.11)
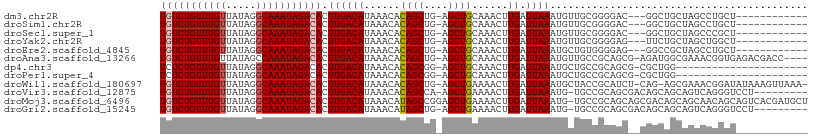

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:16 2011