| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,415,018 – 3,415,113 |
| Length | 95 |
| Max. P | 0.656861 |

| Location | 3,415,018 – 3,415,113 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 77.94 |
| Shannon entropy | 0.44595 |
| G+C content | 0.43837 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -11.72 |
| Energy contribution | -11.69 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 3415018 95 - 23011544 CUGAAUACCAAAAUUGCCCGGCGAGGUUUUCUUAAUUUUUUU-ACUAUGCGAUUUUCGGC-CAUUUGCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((.((((((((((..((..(((............))).-.))..)))))))).)).-........((((((.....)))))))))))...... ( -20.70, z-score = -0.99, R) >droPer1.super_1 2654092 89 + 10282868 CUGAAUACCAAAAAUGCCUGUUGGGGGUU-------UUUUUCCACCGUUUUUUUUUCGGA-CACUUUUAGCCACUUAAGUAGUGGCAUUCACCUCCG ......................((((((.-------........(((.........))).-........((((((.....))))))....)))))). ( -22.00, z-score = -1.67, R) >dp4.chr4_group3 5556257 90 + 11692001 CUGAAUACCAAAAAUGCCUGUUGGGGGUUU------UUUUCCACCGUUUUUUUUUUCGGA-CACUUUUAGCCACUUAAGUAGUGGCAUUCACCUCCG ......................((((((..------.......(((..........))).-........((((((.....))))))....)))))). ( -21.90, z-score = -1.70, R) >droAna3.scaffold_12916 2876740 94 + 16180835 CUGAAUACC--AACCGCCUUUUUGGGGGCUCUU-GCUCCAUUCCCAGUCUUAAUGUCGUUGUAUUUUCCGAUUUUCGGGCAGUGACAUUCACCUCCG (((.....(--((........)))(((((....-))))).....)))....(((((((((((.....(((.....))))))))))))))........ ( -20.70, z-score = -0.50, R) >droEre2.scaffold_4929 3444471 96 - 26641161 CUGAAUACCUAAAUUGCCUGGUGAGGUUUUCUUAAUUUUUUUCACUACCCGAUUUUCGGC-CAUUUCCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((.((.((((((..((((((((.............))))))))..))))))..)).-........((((((.....)))))))))))...... ( -22.82, z-score = -2.73, R) >droYak2.chr2L 3400817 95 - 22324452 CUGAAUACAAAAAUUGCCUGGCGAGGUUUUCGUAAUUUUUUU-ACUAUGCGAUUUUCGGC-CAUUUUCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((............((((((((...(((((........-....))))).)))).))-))......((((((.....)))))))))))...... ( -22.20, z-score = -1.70, R) >droSec1.super_5 1561596 95 - 5866729 CUGAAUACCAAAAUUGCCUGGCGAGGUUUUCUUAAUUUUUUU-ACUAUGCGAUUUUCGGC-CAUUUGCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((.((((((((((.(((..(((............))).-.))).)))))))).)).-........((((((.....)))))))))))...... ( -22.70, z-score = -1.64, R) >droSim1.chr2L 3353283 94 - 22036055 CUGAAUACCAAAAUUGCCUGGCGAGGUUUUCUUAA-UUUUUU-ACUAUGCGAUUUUCGGC-CAUUUGCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((.((((((((((.(((.((((((.....))-))))..-.))).)))))))).)).-........((((((.....)))))))))))...... ( -22.80, z-score = -1.71, R) >consensus CUGAAUACCAAAAUUGCCUGGCGAGGUUUUCUUAAUUUUUUU_ACUAUGCGAUUUUCGGC_CAUUUUCAGCCACUCAAGUAGUGGCAUUCACCUCCG .(((((..........(((....)))...........................................((((((.....)))))))))))...... (-11.72 = -11.69 + -0.03)

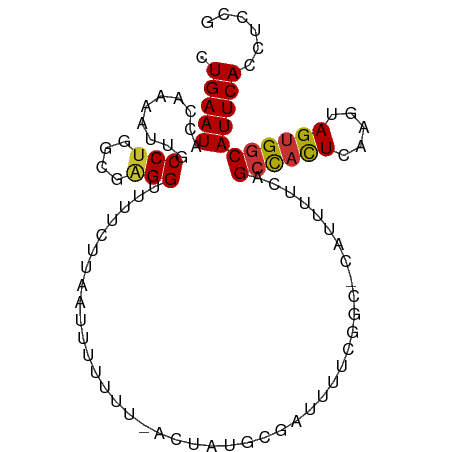

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:50 2011