
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,276,824 – 16,276,934 |
| Length | 110 |
| Max. P | 0.875598 |

| Location | 16,276,824 – 16,276,934 |
|---|---|
| Length | 110 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 87.05 |
| Shannon entropy | 0.25852 |
| G+C content | 0.46494 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -24.65 |
| Energy contribution | -26.10 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16276824 110 + 21146708 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAACCAUAGGGUCCCGCCCCCAACUCACACAUCCA (((.(((......((((((((((((((.(((..-.))).))))))).(((((((....))))))).....))))))).......(((.....))).....))).))).... ( -32.30, z-score = -1.36, R) >droEre2.scaffold_4845 10420778 110 + 22589142 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAUGGGUUAAAAGGCGUAAACAAGCCAUAGGGUCCCGCCCCUUACUCACACAUGCC .(((((.....((.(((.(((((((((.(((..-.))).))))))))).))).))..(((((.....(((........)))...(((.....)))...)))))..))))). ( -34.10, z-score = -1.43, R) >droYak2.chr2R 8214195 110 - 21139217 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAACCAUGGGGUCCCGCCCCUUACACACACAUGCA .(((((.......((((((((((((((.(((..-.))).))))))).(((((((....))))))).....)))))))......((((.....)))).........))))). ( -38.40, z-score = -2.45, R) >droSec1.super_1 13833639 110 + 14215200 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAACCAUAGGGUCCCGCCCCCAACUCACACAUGCA .(((((.......((((((((((((((.(((..-.))).))))))).(((((((....))))))).....))))))).......(((.....)))..........))))). ( -34.90, z-score = -1.84, R) >droSim1.chr2R 14931782 110 + 19596830 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAACCGUAGGGUCCCGCCCCCUACUCACACAUGCA .(((((.......((((((((((((((.(((..-.))).))))))).(((((((....))))))).....)))))))....((((((.......)))))).....))))). ( -39.50, z-score = -2.98, R) >droWil1.scaffold_180697 4115148 95 + 4168966 UGGAUGAUUAAUGUGUUUACACAGGCCACGAGCGUUUGCGGUCUGAGCUGAUUUGAAAGGGUUAAAAGGCGUAAACA------------CAAACACACACACAAACA---- ((..((.....(((((((((.((((((.(((....))).)))))).((((((((....)))))....))))))))))------------))......))..))....---- ( -26.20, z-score = -2.07, R) >droAna3.scaffold_13266 16870079 102 - 19884421 UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC-UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAGCCCUCC----CAUACACACACACAUACA---- .(((((.....(((((...((((((((.(((..-.))).))))))))(((((((....)))))))..(((........)))....----...)))))....))))).---- ( -33.00, z-score = -2.88, R) >consensus UGUAUGAUUAAUGUGUUUACACAGGCCACGAGC_UUUGCGGUCUGUGUUGAUUCGAAAGGGUUAAAAGGCGUAAACAAACCAUAGGGUCCCGCCCCCUACUCACACAUGCA (((.((.......((((((((((((((.(((....))).))))))).(((((((....))))))).....))))))).......(((.....)))......)).))).... (-24.65 = -26.10 + 1.45)

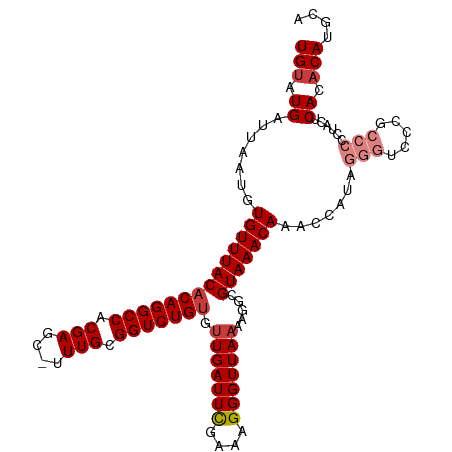
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:13 2011