
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,256,499 – 16,256,576 |
| Length | 77 |
| Max. P | 0.971399 |

| Location | 16,256,499 – 16,256,576 |
|---|---|
| Length | 77 |
| Sequences | 4 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 50.00 |
| Shannon entropy | 0.80583 |
| G+C content | 0.46909 |
| Mean single sequence MFE | -18.21 |
| Consensus MFE | -7.28 |
| Energy contribution | -6.03 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
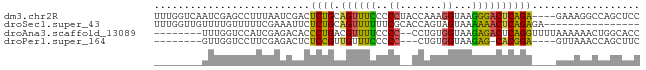
>dm3.chr2R 16256499 77 + 21146708 UUUGGUCAAUCGAGCCUUUAAUCGACUCUGCAGUUUCCCCCUACCAAAGGUAAGGGACUCAGA----GAAAGGCCAGCUCC .((((((..((((........))))(((((.(((...(((.((((...)))).))))))))))----)...)))))).... ( -26.60, z-score = -2.73, R) >droSec1.super_43 56432 65 + 327307 UUUGGUUGUUUUGUUUUUCGAAAUUCUCUGCAGUUUUUUCGCACCAGUAGUAAAAAACUCAGAGA---------------- .......((((((.....))))))((((((.((((((((.((.......))))))))))))))))---------------- ( -15.10, z-score = -2.19, R) >droAna3.scaffold_13089 423646 71 - 978470 --------UUUGGUCCAUCGAGACACCCUGACGUUUUCCCC--CCUGUGGUAAGAGACUCAGGUUUUAAAAAACUGGCACC --------...((((((.........(((((.(((((...(--(....))...))))))))))...........))).))) ( -13.95, z-score = 0.64, R) >droPer1.super_164 85937 65 + 94152 --------GUUGGUCCUUCGAGACUCUCCGUUGUUUCCCCC---CUGUGGUAAGAG-CACGGA----GUUAAACCAGCUUC --------((((((.(((((.(.(((((((..(........---)..)))..))))-).))))----)....))))))... ( -17.20, z-score = -1.02, R) >consensus ________UUUGGUCCUUCGAGACACUCUGCAGUUUCCCCC__CCAGUGGUAAGAGACUCAGA____GAAAAACCAGCUCC ..........................((((.((((((.((........))...)))))))))).................. ( -7.28 = -6.03 + -1.25)
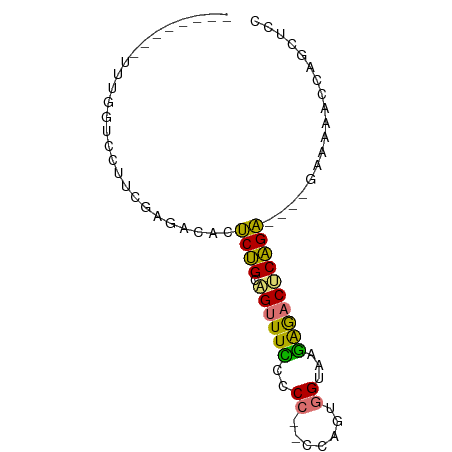
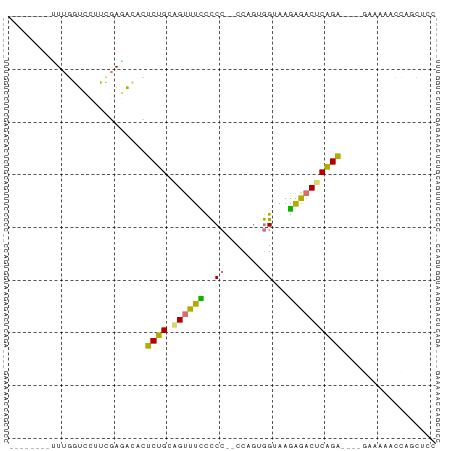
| Location | 16,256,499 – 16,256,576 |
|---|---|
| Length | 77 |
| Sequences | 4 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 50.00 |
| Shannon entropy | 0.80583 |
| G+C content | 0.46909 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -9.49 |
| Energy contribution | -9.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.71 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16256499 77 - 21146708 GGAGCUGGCCUUUC----UCUGAGUCCCUUACCUUUGGUAGGGGGAAACUGCAGAGUCGAUUAAAGGCUCGAUUGACCAAA ((..(.((((((((----(((((((((((((((...))))))))...))).)))))......))))))).).....))... ( -27.40, z-score = -1.52, R) >droSec1.super_43 56432 65 - 327307 ----------------UCUCUGAGUUUUUUACUACUGGUGCGAAAAAACUGCAGAGAAUUUCGAAAAACAAAACAACCAAA ----------------((((((((((((((..((....))..)))))))).))))))........................ ( -13.30, z-score = -2.39, R) >droAna3.scaffold_13089 423646 71 + 978470 GGUGCCAGUUUUUUAAAACCUGAGUCUCUUACCACAGG--GGGGAAAACGUCAGGGUGUCUCGAUGGACCAAA-------- (((.(((......((...(((((((.(((..((...))--..)))..)).))))).))......))))))...-------- ( -18.30, z-score = 0.38, R) >droPer1.super_164 85937 65 - 94152 GAAGCUGGUUUAAC----UCCGUG-CUCUUACCACAG---GGGGGAAACAACGGAGAGUCUCGAAGGACCAAC-------- .....(((.....(----(((((.-(((((.....))---)))(....).))))))....((....)))))..-------- ( -17.20, z-score = -0.13, R) >consensus GGAGCUGGUUUUUC____UCUGAGUCUCUUACCACAGG__GGGGAAAACUGCAGAGAGUCUCGAAGGACCAAA________ ..................(((((.(((((((((...))))))))).....))))).......................... ( -9.49 = -9.55 + 0.06)


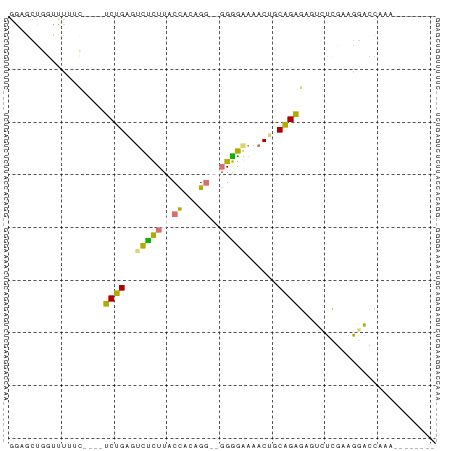
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:10 2011