
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,252,232 – 16,252,358 |
| Length | 126 |
| Max. P | 0.659942 |

| Location | 16,252,232 – 16,252,358 |
|---|---|
| Length | 126 |
| Sequences | 9 |
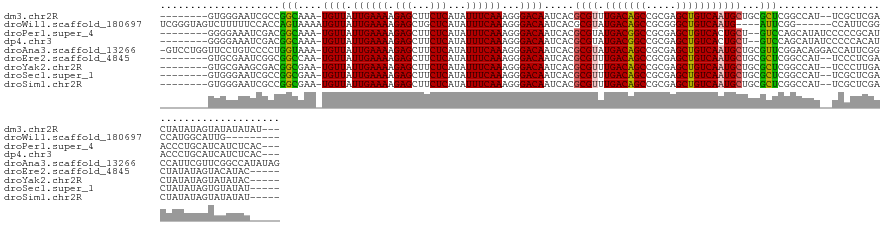
| Columns | 140 |
| Reading direction | forward |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.49137 |
| G+C content | 0.48519 |
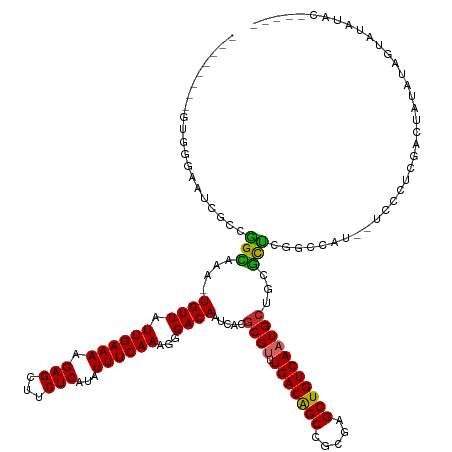
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.659942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16252232 126 + 21146708 --------GUGGGAAUCGCCGGCAAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU--UCGCUCGACUAUAUAGUAUAUAUAU--- --------(.(.((((.((((((...-((((.((((((.(((...)))...))))))...)))).....(((((.((((((.....)))))))))))...)).)))).))--)).).)...((((((...)))))).--- ( -36.80, z-score = -1.11, R) >droWil1.scaffold_180697 11086 121 - 4168966 UCGGGUAGUCUUUUUCCACCAGUAAAAUGUUAUUGAAAAGAGCUGCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACAGCCGCGGGCUGUCAAUG----AUUCGG------CCAUUCGGCCAUGGCAUUG--------- ..((((((.((((((....((((((....)))))).)))))))))))).....................((...((((((((...))))))))...----....((------((....))))...))....--------- ( -36.50, z-score = -1.62, R) >droPer1.super_4 5650767 126 + 7162766 --------GGGGAAAUCGACGGCAAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACGGCCGCGAGCUGUCACUGCU--GUCCAGCAUAUCCCCCGCAUACCCUGCAUCAUCUCAC--- --------(((((....((((((...-((((.((((((.(((...)))...))))))...))))....((((.........)))).))))))..((((--....))))..))))).(((.....)))..........--- ( -35.70, z-score = -1.17, R) >dp4.chr3 10316937 126 + 19779522 --------GGGGAAAUCGACGGCAAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACGGCCGCGAGCUGUCACUGCU--GUCCAGCAUAUCCCCCACAUACCCUGCAUCAUCUCAC--- --------(((((....((((((...-((((.((((((.(((...)))...))))))...))))....((((.........)))).))))))..((((--....))))..)))))......................--- ( -33.50, z-score = -0.91, R) >droAna3.scaffold_13266 16854245 138 - 19884421 -GUCCUGGUUCCUGUCCCCUGGUAAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUAUGACAGCCGCGAGCUGUCAAUGCUGCGUUCGGACAGGACCAUUCGGCCAUUCGUUCGGCCAUAUAG -((((((..(((((((((...(.(((-(((....(((......)))..)))))))...))))))...((((((((((((((.....)))))).)))).))))..))))))))).....((((........))))...... ( -49.20, z-score = -3.45, R) >droEre2.scaffold_4845 10405212 124 + 22589142 --------GUGCGAAUCGGCGGCCAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU--UCCCUCGACUAUAUAGUACAUAC----- --------(((((......)((((..-((((.((((((.(((...)))...))))))...)))).....((((((((((((.....))))))))....))))..))))..--...............))))....----- ( -33.60, z-score = -0.33, R) >droYak2.chr2R 8198304 124 - 21139217 --------GUGCGAAGCGACGGCGAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU--UCCCUUGACUAUAUAGUAUAUAC----- --------(((((((((.....(((.-.....)))......)))))..((((....((((((.......(((((.((((((.....))))))))))).((.....))...--))))))....)))).))))....----- ( -33.80, z-score = -0.40, R) >droSec1.super_1 13818329 124 + 14215200 --------GUGGGAAUCGCCGGCGAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU--UCGCUCGACUAUAUAGUGUAUAU----- --------(.(.((((.((((((((.-((((.((((((.(((...)))...))))))...)))).))..(((((.((((((.....)))))))))))...)).)))).))--)).).).................----- ( -38.80, z-score = -1.07, R) >droSim1.chr2R 14916427 124 + 19596830 --------GUGGGAAUCGCCGGCGAA-UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU--UCGCUCGACUAUAUAGUAUAUAU----- --------(.(.((((.((((((((.-((((.((((((.(((...)))...))))))...)))).))..(((((.((((((.....)))))))))))...)).)))).))--)).).).................----- ( -38.80, z-score = -1.37, R) >consensus ________GUGGGAAUCGCCGGCAAA_UGUUAUUGAAAAGAGCUUCUCAUAUUUCAAAGGGACAAUCACGCGUUUGACAGCCGCGAGCUGUCAAUGCUGCGCUCGGCCAU__UCCCUCGACUAUAUAGUAUAUAC_____ ....................(((....((((.((((((.(((...)))...))))))...)))).....((((.(((((((.....)))))))))))...)))..................................... (-20.71 = -20.08 + -0.63)
| Location | 16,252,232 – 16,252,358 |
|---|---|
| Length | 126 |
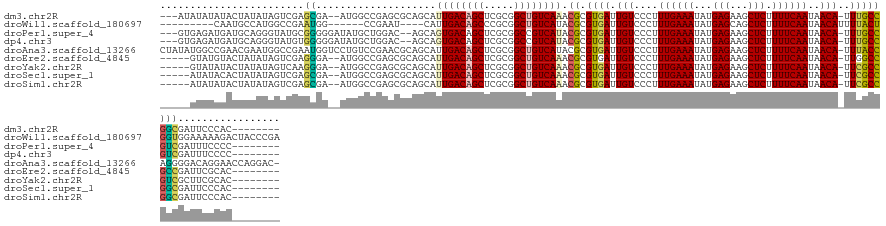
| Sequences | 9 |
| Columns | 140 |
| Reading direction | reverse |
| Mean pairwise identity | 75.54 |
| Shannon entropy | 0.49137 |
| G+C content | 0.48519 |
| Mean single sequence MFE | -37.74 |
| Consensus MFE | -19.28 |
| Energy contribution | -18.58 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16252232 126 - 21146708 ---AUAUAUAUACUAUAUAGUCGAGCGA--AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUUGCCGGCGAUUCCCAC-------- ---.((((((...))))))((((.((((--(((..(((.(((.((.((((((((.....))))))))..))))).))).....((((((...(((...))).))))))...))-))))))))).........-------- ( -36.80, z-score = -1.79, R) >droWil1.scaffold_180697 11086 121 + 4168966 ---------CAAUGCCAUGGCCGAAUGG------CCGAAU----CAUUGACAGCCCGCGGCUGUCAUACGCGUGAUUGUCCCUUUGAAAUAUGAGCAGCUCUUUUCAAUAACAUUUUACUGGUGGAAAAAGACUACCCGA ---------....((..(((((....))------)))...----...((((((((...))))))))...))((((.(((....((((((...(((...))).))))))..)))..)))).(((((.......)))))... ( -33.80, z-score = -1.61, R) >droPer1.super_4 5650767 126 - 7162766 ---GUGAGAUGAUGCAGGGUAUGCGGGGGAUAUGCUGGAC--AGCAGUGACAGCUCGCGGCCGUCAUACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUUGCCGUCGAUUUCCCC-------- ---..(((((((((((((((.....(((((((((((....--))))(((((.((.....)).))))).........)))))))((((((...(((...))).))))))..)).-)))).)))).)))))...-------- ( -34.00, z-score = 0.92, R) >dp4.chr3 10316937 126 - 19779522 ---GUGAGAUGAUGCAGGGUAUGUGGGGGAUAUGCUGGAC--AGCAGUGACAGCUCGCGGCCGUCAUACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUUGCCGUCGAUUUCCCC-------- ---..((((((((....((((.((((((((((((((....--))))(((((.((.....)).))))).........)))))))((((((...(((...))).))))))...))-).))))))).)))))...-------- ( -35.10, z-score = 0.45, R) >droAna3.scaffold_13266 16854245 138 + 19884421 CUAUAUGGCCGAACGAAUGGCCGAAUGGUCCUGUCCGAACGCAGCAUUGACAGCUCGCGGCUGUCAUACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUUACCAGGGGACAGGAACCAGGAC- ((((.((((((......)))))).))))((((((((..((((.....(((((((.....)))))))...))))...((((((((.(((((.((((((....)))))).....)-))).).)))))))))))..))))).- ( -50.20, z-score = -3.71, R) >droEre2.scaffold_4845 10405212 124 - 22589142 -----GUAUGUACUAUAUAGUCGAGGGA--AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUGGCCGCCGAUUCGCAC-------- -----..............(.((((...--.(((((((((((....((((((((.....))))))))..))))...(((....((((((...(((...))).))))))..)))-)))))))....)))))..-------- ( -35.80, z-score = -0.39, R) >droYak2.chr2R 8198304 124 + 21139217 -----GUAUAUACUAUAUAGUCAAGGGA--AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUCGCCGUCGCUUCGCAC-------- -----..............((((.....--.))))...(((.(((.((((((((.....))))))))(((.((((.(((....((((((...(((...))).))))))..)))-.))))))).))).)))..-------- ( -37.20, z-score = -1.48, R) >droSec1.super_1 13818329 124 - 14215200 -----AUAUACACUAUAUAGUCGAGCGA--AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUCGCCGGCGAUUCCCAC-------- -----..............((((.((((--(((..(((.(((.((.((((((((.....))))))))..))))).))).....((((((...(((...))).))))))...))-))))))))).........-------- ( -38.40, z-score = -2.15, R) >droSim1.chr2R 14916427 124 - 19596830 -----AUAUAUACUAUAUAGUCGAGCGA--AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA-UUCGCCGGCGAUUCCCAC-------- -----..............((((.((((--(((..(((.(((.((.((((((((.....))))))))..))))).))).....((((((...(((...))).))))))...))-))))))))).........-------- ( -38.40, z-score = -2.15, R) >consensus _____GUAUAUACUAUAUAGUCGAGGGA__AUGGCCGAGCGCAGCAUUGACAGCUCGCGGCUGUCAAACGCGUGAUUGUCCCUUUGAAAUAUGAGAAGCUCUUUUCAAUAACA_UUUGCCGGCGAUUCCCAC________ ........................((....................((((((((.....)))))))).((.((((........((((((...(((...))).)))))).......))))))))................. (-19.28 = -18.58 + -0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:09 2011