| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,251,600 – 16,251,694 |
| Length | 94 |
| Max. P | 0.765647 |

| Location | 16,251,600 – 16,251,694 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 63.81 |
| Shannon entropy | 0.68029 |
| G+C content | 0.59255 |
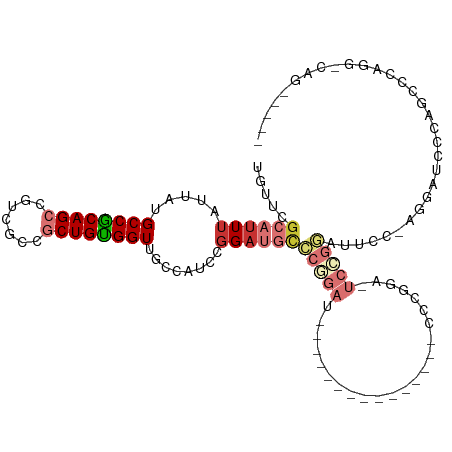
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -12.44 |
| Energy contribution | -13.19 |
| Covariance contribution | 0.75 |
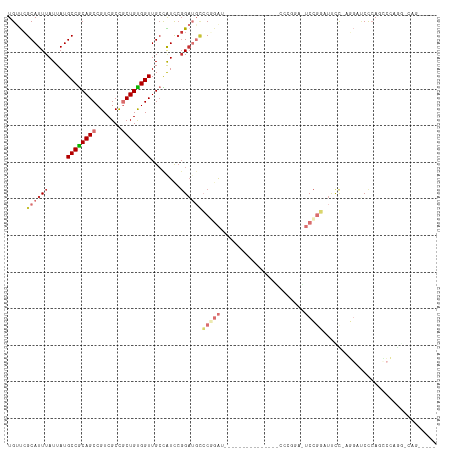
| Combinations/Pair | 1.21 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
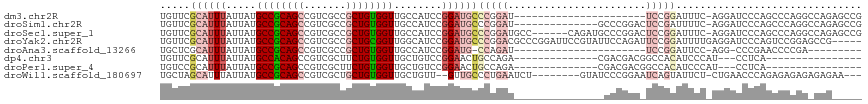
>dm3.chr2R 16251600 94 - 21146708 UGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAU----------------------UCCGGAUUUC-AGGAUCCCAGCCCAGGCCAGAGCCG .((((...........((((((((.......)))))))).(((((((((....))))))----------------------...(((((..-..))))).......)))..)))).. ( -33.00, z-score = -1.03, R) >droSim1.chr2R 14915845 102 - 19596830 UGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAU--------------GCCCGGACUCCGAUUUUC-AGGAUCCCAGCCCAGGCCAGAGCCG .((((...........((((((((.......)))))))).(((((((((....))))))--------------((..(((.(((.......-.))))))..))...)))..)))).. ( -37.50, z-score = -1.50, R) >droSec1.super_1 13817712 110 - 14215200 UGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAUGCC------CAGAUGCCCGGACUCCGGAUUUC-AGGAUCCCAGCCCAGGCCAGAGCCG .....((((((.....((((((((.......))))))))...(((((((....)))))))..------.)))))).(((.(((.(((((..-..)))))..((....))..)))))) ( -41.20, z-score = -1.28, R) >droYak2.chr2R 8197596 112 + 21139217 UGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGCGGUUGGCAUCCGGAUGCCCGGACGCCCGGAUUCCGUAUUCCAGAUUCCGGAUUUUGAGGAUCCCAGUCCGGAGCCG----- ................((((((((.......)))))))).(((.(((((((....(((..((((((.((((.(........).)))).)))).)).)))..)))))))))).----- ( -43.90, z-score = -1.63, R) >droAna3.scaffold_13266 16853873 83 + 19884421 UGCUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUG-CCAGAU----------------------UCCGGAUUCC-AGG-CCCGAACCCCGA--------- ...(((..........((((((((.......)))))))).((((((((((..-......----------------------)))))))...-.))-).))).......--------- ( -27.20, z-score = -0.81, R) >dp4.chr3 10316660 84 - 19779522 UGUUCGCAUUUAUUAUGCCACAGCCGUCGCUUCUGUGGUUGCUGUCCGGAACUGCCAGA--------------CGACGACGGCCACAUCCCAU---CCUCA---------------- (((..((((.....)))).)))((((((((.((((..(((.(.....).)))..).)))--------------.).)))))))..........---.....---------------- ( -26.40, z-score = -1.89, R) >droPer1.super_4 5650497 84 - 7162766 UGUCCGCAUUUAUUAUGCCGCAGCCGUCGCUUCUGUGGUUGCUGUCCGGAACUGCCAGA--------------CGACGACGGCCACAUCCCAU---CCUCA---------------- (((..((((.....)))).)))((((((((.((((..(((.(.....).)))..).)))--------------.).)))))))..........---.....---------------- ( -26.10, z-score = -1.29, R) >droWil1.scaffold_180697 10153 103 + 4168966 UGCUAGCAUUUAUUAUGCCGCAGCCGUCGCUGCUGUGGUUGCUGUU--GUUGCCCUGAAUCU--------GUAUCCCGGAAUCAGUAUUCU-CUGAACCCAGAGAGAGAGAGAA--- .((.((((........((((((((.......))))))))......)--))))).((((.(((--------(.....)))).))))..((((-(((....)))))))........--- ( -31.34, z-score = -1.87, R) >consensus UGUUCGCAUUUAUUAUGCCGCAGCCGUCGCCGCUGUGGUUGCCAUCCGGAUGCCCGGAU_______________CCCGGA_UCCGGAUUCC_AGGAUCCCAGCCCAGG_CAG_____ .....((((((.....((((((((.......))))))))........))))))................................................................ (-12.44 = -13.19 + 0.75)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:07 2011