| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,249,121 – 16,249,212 |
| Length | 91 |
| Max. P | 0.937235 |

| Location | 16,249,121 – 16,249,212 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.73706 |
| G+C content | 0.45933 |
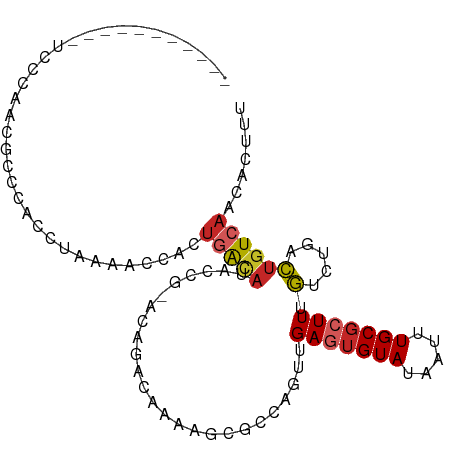
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -4.73 |
| Energy contribution | -5.06 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.937235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
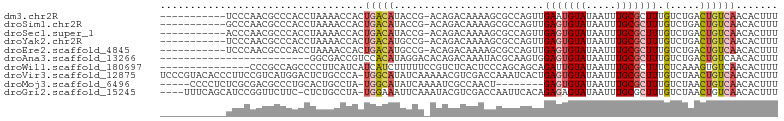
>dm3.chr2R 16249121 91 + 21146708 -----------UCCCAACGCCCACCUAAAACCACUGACAUACCG-ACAGACAAAAGCGCCAGUUGAAUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -----------.......................(((((....(-.((((((((.((((.(((((......))))).)))))))))))).))))))....... ( -21.70, z-score = -3.96, R) >droSim1.chr2R 14913474 91 + 19596830 -----------GCCCAACGCCCACCUAAAACCACUGACAUACCG-ACAGACAAAAGCGCCAGUUGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -----------.......................(((((....(-.((((((((.((((.(((((......))))).)))))))))))).))))))....... ( -21.70, z-score = -2.88, R) >droSec1.super_1 13815264 91 + 14215200 -----------ACCCAACGCCCACCUAAAACCACUGACAUACCG-ACAGACAAAAGCGCCAGUUGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -----------.......................(((((....(-.((((((((.((((.(((((......))))).)))))))))))).))))))....... ( -21.70, z-score = -3.16, R) >droYak2.chr2R 8195146 91 - 21139217 -----------UCCCAACGCCCACCUAAAACCACUGACAUGCCG-ACAGACAAAAGCGCCAGUUGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -----------.......................(((((....(-.((((((((.((((.(((((......))))).)))))))))))).))))))....... ( -21.70, z-score = -2.60, R) >droEre2.scaffold_4845 10402076 91 + 22589142 -----------UCCCAACGCCCACCUAAAACCACUGACAUGCCG-ACAGACAAAAGCGCCAGUUGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -----------.......................(((((....(-.((((((((.((((.(((((......))))).)))))))))))).))))))....... ( -21.70, z-score = -2.60, R) >droAna3.scaffold_13266 16851408 78 - 19884421 -------------------------GGCGACCGUCCACAUAGGACACAGACAAAUACGCAAGUGGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU -------------------------((((...((((.....)))).((((((..(((....)))(((((((.....)))))))))))))..))))........ ( -26.80, z-score = -3.43, R) >droWil1.scaffold_180697 5757 88 - 4168966 ---------------CCCGCCAGCCCCUUCAUCAUCAUCUUUUUCCGUCUCACUCCCAGCAGCAGAUUGUAUAAUUUGCGCUUUCUCAAAGUGUCAACACUUU ---------------..........................................(((.(((((((....)))))))))).....((((((....)))))) ( -12.30, z-score = -2.37, R) >droVir3.scaffold_12875 11622577 102 - 20611582 UCCCGUACACCCUUCCGUCAUGGACUCUGCCCA-UGGCAUAUCAAAAACGUCGACCAAAUCACUGAGUGUAUAAUUUGCGCUUUGUCUAACUGUCAACACUUU ................(((((((.......)))-))))...........((.(((......((.(((((((.....))))))).))......))).))..... ( -21.90, z-score = -2.18, R) >droMoj3.scaffold_6496 18191601 89 + 26866924 -----CCCCUCUCGCGACGCCCUGCACUGCCUA-UGGCAUAUCAAAAUCGCCAACU--------GAGUGUAUAAUUUGCGCUUUGUCUAACUGUCAACACUUU -----........((((.((...))..((((..-.))))........))))..((.--------(((((((.....))))))).))................. ( -15.30, z-score = -0.68, R) >droGri2.scaffold_15245 15992136 97 - 18325388 ----UUUCAGCAUCCGGUUCUUC-CUCUGCCUA-UGGAAAUUCAAAUACGUCGACCAAUUCACAGAGAGUAUAAUUUGCGCUUUGUCUAACUGUCAACACUUU ----((((((((...((.....)-)..)))...-)))))..........((.(((......((((((.(((.....))).))))))......))).))..... ( -15.80, z-score = -0.31, R) >consensus ___________UCCCAACGCCCACCUAAAACCACUGACAUACCG_ACAGACAAAAGCGCCAGUUGAGUGUAUAAUUUGCGCUUUGUCUGACUGUCAACACUUU ..................................(((((.........................(((((((.....))))))).(.....))))))....... ( -4.73 = -5.06 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:06 2011