| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,414,581 – 3,414,716 |
| Length | 135 |
| Max. P | 0.979598 |

| Location | 3,414,581 – 3,414,693 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.33 |
| Shannon entropy | 0.48287 |
| G+C content | 0.36585 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -12.75 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
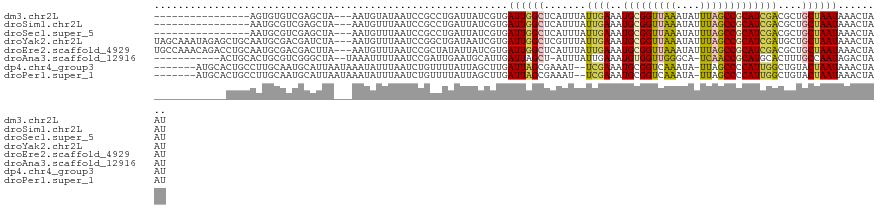
>dm3.chr2L 3414581 112 - 23011544 UCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAUUAAAACAUUGCU-CACUGAAAUGAAUAAACACC------- ..............((((((((((((.....))).((((((((((....)))))))))).......)))))).............((((.(.-....).))))......))).------- ( -21.70, z-score = -1.61, R) >droSim1.chr2L 3352831 112 - 22036055 UCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAUUAAAACAUUGCC-CACUGAAAUGAAUAAACACC------- ..............((((((((((((.....))).((((((((((....)))))))))).......)))))).............((((...-......))))......))).------- ( -21.40, z-score = -1.59, R) >droSec1.super_5 1561136 112 - 5866729 UCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAUUAAAACAUUGCC-CACUGAAAUGAAUAAACACC------- ..............((((((((((((.....))).((((((((((....)))))))))).......)))))).............((((...-......))))......))).------- ( -21.40, z-score = -1.59, R) >droYak2.chr2L 3400362 113 - 22324452 UCCGGCUGAUAAUCGUGAUUGGCUCGUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGAUGCUGCUAAUAAACUAAUUAAAACAUUGCCGCGCUGAAAUGAAUAAACACC------- ...(((..((.......))..))).(((((((.(..(((((((((....)))))))))(((.(((.((.....................)).))).)))..).)))))))...------- ( -27.20, z-score = -1.83, R) >droEre2.scaffold_4929 3444023 112 - 26641161 UCCGCUAUAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAUUAAAACAUUGCU-CACUGAAAUGCAUAAACACC------- ..............((((((((((((.....))).((((((((((....)))))))))).......))))))................(((.-.........)))....))).------- ( -22.00, z-score = -1.39, R) >droAna3.scaffold_12916 2876418 97 + 16180835 UCCGAUUGAAUGCAUUGAUUAGCU-AUUUAUUGAAAUGUGGUUGGGCA-UCAACCGCAUGCACUUUGCCAAUAGACUAAUUAAAACUCGGAUGAACUCC--------------------- (((((.((....))((((((((..-.(((((((..(((((((((....-.)))))))))((.....)))))))))))))))))...)))))........--------------------- ( -26.00, z-score = -2.76, R) >dp4.chr4_group3 5554559 116 + 11692001 UCUGUUUUAUUAGCUUGAUUAGCGAAAUU--CGAAAUGCGGUCAAAUA-UUAGCCCCAUUGGCUGUACUAAUAAACUAAUUAAAACAGUGCC-AAAUAUAAUGGAUACGUUUCCAAAACC .((((((((((((....(((((((.....--))...(((((((((...-.........))))))))))))))...)))).))))))))....-........((((......))))..... ( -24.30, z-score = -2.13, R) >droPer1.super_1 2652308 116 + 10282868 UCUGUUUUAUUAGCUUGAUUAGCGAAAUU--CGAAAUGCGGUCAAAUA-UUAGCCCCAUUGGCUGUACUAAUAAACUAAUUAAAACAGUGCC-AAAUAUAAUGGAUACGUUUCCAAAACC .((((((((((((....(((((((.....--))...(((((((((...-.........))))))))))))))...)))).))))))))....-........((((......))))..... ( -24.30, z-score = -2.13, R) >consensus UCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAUUAAAACAUUGCC_CACUGAAAUGAAUAAACACC_______ ...............(((((((.............((((((((((....))))))))))................)))))))...................................... (-12.75 = -12.50 + -0.25)


| Location | 3,414,613 – 3,414,716 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 122 |
| Reading direction | reverse |
| Mean pairwise identity | 73.07 |
| Shannon entropy | 0.51366 |
| G+C content | 0.36595 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -13.19 |
| Energy contribution | -13.15 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979598 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 3414613 103 - 23011544 ----------------AGUGUGUCGAGCUA---AAUGUAUAAUCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAU ----------------((((((((((((((---((((.((((((.....)))))))))..)))))))..........((((((((((....)))))))))).))))).)))........... ( -34.00, z-score = -4.47, R) >droSim1.chr2L 3352863 103 - 22036055 ----------------AAUGCGUCGAGCUA---AAUGUUUAAUCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAU ----------------...(((((((((((---(((((((((.(((((..(((.....)))..).(((.....)))...)))))))))))))))))....)))))))............... ( -34.00, z-score = -4.47, R) >droSec1.super_5 1561168 103 - 5866729 ----------------AAUGCGUCGAGCUA---AAUGUUUAAUCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAU ----------------...(((((((((((---(((((((((.(((((..(((.....)))..).(((.....)))...)))))))))))))))))....)))))))............... ( -34.00, z-score = -4.47, R) >droYak2.chr2L 3400395 119 - 22324452 UAGCAAAUAGAGCUGCAAUGCGACGAUCUA---AAUGUUUAAUCCGGCUGAUAAUCGUGAUUGGCUCGUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGAUGCUGCUAAUAAACUAAU (((((....((((..((((((((..(((..---...(((......))).)))..)))).))))))))...((((((..(((((((((....))))))))))))))).))))).......... ( -32.50, z-score = -2.02, R) >droEre2.scaffold_4929 3444055 119 - 26641161 UGCCAAACAGACCUGCAAUGCGACGACUUA---AAUGUUUAAUCCGCUAUAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAU ..............(((..(((.(((..((---((((.((((((((.........)).))))))..))))))......(((((((((....)))))))))))).))))))............ ( -26.90, z-score = -1.58, R) >droAna3.scaffold_12916 2876437 107 + 16180835 -----------ACUGCACUGCGUCGGGCUA--UAAAUUUUAAUCCGAUUGAAUGCAUUGAUUAGCU-AUUUAUUGAAAUGUGGUUGGGCA-UCAACCGCAUGCACUUUGCCAAUAGACUAAU -----------..((((....(((((..((--.......))..)))))....))))...(((((..-.(((((((..(((((((((....-.)))))))))((.....)))))))))))))) ( -28.70, z-score = -1.78, R) >dp4.chr4_group3 5554598 112 + 11692001 -------AUGCACUGCCUUGCAAUGCAUUAAUAAAUAUUUAAUCUGUUUUAUUAGCUUGAUUAGCGAAAU--UCGAAAUGCGGUCAAAUA-UUAGCCCCAUUGGCUGUACUAAUAAACUAAU -------(((((.(((...))).)))))...................((((((((.((((((.(((....--......)))))))))...-.(((((.....)))))..))))))))..... ( -20.10, z-score = -0.26, R) >droPer1.super_1 2652347 112 + 10282868 -------AUGCACUGCCUUGCAAUGCAUUAAUAAAUAUUUAAUCUGUUUUAUUAGCUUGAUUAGCGAAAU--UCGAAAUGCGGUCAAAUA-UUAGCCCCAUUGGCUGUACUAAUAAACUAAU -------(((((.(((...))).)))))...................((((((((.((((((.(((....--......)))))))))...-.(((((.....)))))..))))))))..... ( -20.10, z-score = -0.26, R) >consensus ____________CUGCAAUGCGACGAGCUA___AAUGUUUAAUCCGCCUGAUUAUCGUGAUUGGCUCAUUUAUUGAAAUGCGGUUAAAUAUUUAGCCGCAUCGACGCUGCUAAUAAACUAAU ...........................................................((((((.......((((..(((((((((....)))))))))))))....))))))........ (-13.19 = -13.15 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:49 2011