
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,223,291 – 16,223,382 |
| Length | 91 |
| Max. P | 0.566689 |

| Location | 16,223,291 – 16,223,382 |
|---|---|
| Length | 91 |
| Sequences | 7 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 82.42 |
| Shannon entropy | 0.36272 |
| G+C content | 0.37801 |
| Mean single sequence MFE | -19.29 |
| Consensus MFE | -11.24 |
| Energy contribution | -12.49 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.566689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
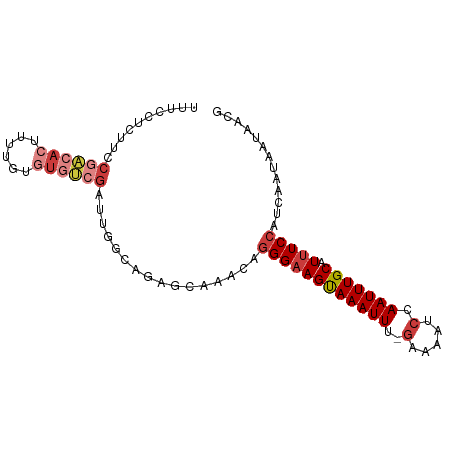
>dm3.chr2R 16223291 91 - 21146708 UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAGAGCAAACAGGGAAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ....((((.(((((((......)))))....)).))))......((((((((((((.-(.....).))))))).)))))............. ( -21.30, z-score = -2.18, R) >droSim1.chr2R 14888249 91 - 19596830 UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAGAGCAAACAGGGAAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ....((((.(((((((......)))))....)).))))......((((((((((((.-(.....).))))))).)))))............. ( -21.30, z-score = -2.18, R) >droSec1.super_1 13789516 91 - 14215200 UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAGAGCAAACAGGGAAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ....((((.(((((((......)))))....)).))))......((((((((((((.-(.....).))))))).)))))............. ( -21.30, z-score = -2.18, R) >droYak2.chr2R 8161556 91 + 21139217 UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAGAGCAAACAGGGAAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ....((((.(((((((......)))))....)).))))......((((((((((((.-(.....).))))))).)))))............. ( -21.30, z-score = -2.18, R) >droEre2.scaffold_4845 10376163 91 - 22589142 UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAAAGCAAACAGGGAAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ..........((((((......))))))((((((...)).....((((((((((((.-(.....).))))))).)))))..))))....... ( -20.10, z-score = -2.16, R) >droAna3.scaffold_13266 16827260 77 + 19884421 --------------UCCUGCUCUCCUCUAUUGGCAAAGCAAACAGGGUAGUAAAUUU-GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG --------------...((((..((......))...))))....(((..(((((((.-(.....).)))))))...)))............. ( -12.00, z-score = -1.20, R) >droVir3.scaffold_12875 11583186 87 + 20611582 UGUCUGGGGCCUGCAGUGCCUGAUCCUGAGAGCAAAACCAAACAAGGAAGCAAAUUUCCAAAAUCCAAUUUGCAUUUCCAUCAAUAA----- (((.(((.((((.(((.........))))).))....))).))).(((((((((((..........)))))))..))))........----- ( -17.70, z-score = 0.15, R) >consensus UUUCCUCUUCCGACACUUUUGUGUGUCGAUUGGCAGAGCAAACAGGGAAGUAAAUUU_GAAAAUCCAAUUUGCAUUUCCAUCAAUAAUAACG ..........((((((......))))))................((((((((((((..........))))))).)))))............. (-11.24 = -12.49 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:02 2011