| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,202,395 – 16,202,485 |
| Length | 90 |
| Max. P | 0.551399 |

| Location | 16,202,395 – 16,202,485 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.85 |
| Shannon entropy | 0.42706 |
| G+C content | 0.39915 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -8.93 |
| Energy contribution | -9.30 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.551399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
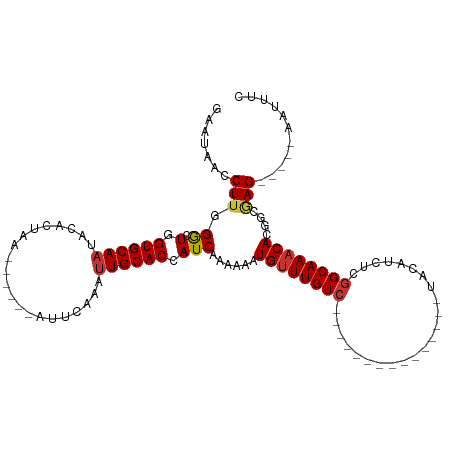
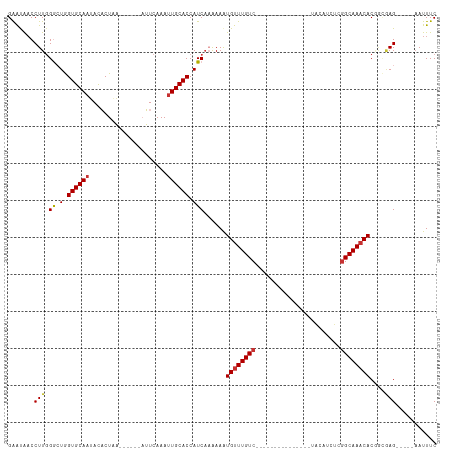
>dm3.chr2R 16202395 90 - 21146708 GAAUAACCUUGGGCUGGUGCAAUACACUAA------AUUCAAAUUGCACCAUCAAAAAAUGUUUGUC---------------UACAUCUCGGCAAACACGGCGAG-----AAUUUC .......((((((.(((((((((.......------......)))))))))))......((((((((---------------........))))))))...))))-----...... ( -23.12, z-score = -3.31, R) >droPer1.super_4 5176340 109 - 7162766 GGACGUCCUUGGG-UAGUGCAAAAUUCUCU------CAUCAAAUUGCACCAUCACAAAAUGUUUGUCUUUAUUUGUACAAUGUGCAUCUCGGCAAACACCAGAAGAGAACAAUUUC ((......(((((-(.((((((........------.......)))))).))).)))..((((((((......(((((...)))))....))))))))))................ ( -21.86, z-score = -0.55, R) >dp4.chr3 9858904 109 - 19779522 GGACGACCUUGGG-UAGUGCAAAAUUCUCU------CAUCAAAUUGCACCAUCACAAAAUGUUUGUCUACAUUUGUACAAUGUGCAUCUUUGCAAACACCAGAAGAGAACAAUUUC ((......(((((-(.((((((........------.......)))))).))).)))..(((((((.......(((((...))))).....)))))))))................ ( -19.46, z-score = 0.30, R) >droAna3.scaffold_13266 16803497 97 + 19884421 GAGCGACCUU-GACUAGUGCAAAAUACUUA------AUUCAAAUUGCACCAUCAAGAAGUGCUUGUCAACG-------GCAUUACAUCCCGGCAAACACACUAAG-----AAUUUC .......(((-((...((((((........------.......))))))..)))))..(((.(((((...(-------(........)).))))).)))......-----...... ( -17.56, z-score = -0.96, R) >droEre2.scaffold_4845 10353606 89 - 22589142 GAAUAACCUUGGGCUGGUGCAAUACACUAA------AUUCAAAUUGCACCAUC-AAAAAUGUUUGUC---------------UACAUCUCGGCAAACACGGCGAG-----AAUUUC .......((((((.(((((((((.......------......)))))))))))-.....((((((((---------------........))))))))...))))-----...... ( -23.12, z-score = -3.26, R) >droYak2.chr2R 8138665 96 + 21139217 GAAUAACCUUGGACUGGUGCAUUACACUAAAUUCAAAUUCAAAUUGCACCAUCAAAAAAUGUUUGUC---------------UACAUCUCGGCAAACACGGCCAG-----CAUUUC ........((((.((((((((.......................)))))).........((((((((---------------........)))))))).))))))-----...... ( -20.50, z-score = -2.56, R) >droSec1.super_1 13768219 90 - 14215200 GAAUAACCUUGGGCUGGUGCAAUACACUAA------AUUCAAAUUGCACCAUCAAAAAAUGUUUGUC---------------UGCAACUCGGCAAACACGGCGAG-----AAUUUC .......((((((.(((((((((.......------......)))))))))))......((((((((---------------........))))))))...))))-----...... ( -23.12, z-score = -2.72, R) >droSim1.chr2R 14861203 90 - 19596830 GAAUAACCUUGGGCUGGUGCAAUACACUAA------AUUCAAAUUGCACCAUCAAAAAAUGUUUGUC---------------UACAACUCGGCAAACACGGCGAG-----AAUUUC .......((((((.(((((((((.......------......)))))))))))......((((((((---------------........))))))))...))))-----...... ( -23.12, z-score = -3.36, R) >consensus GAAUAACCUUGGGCUGGUGCAAUACACUAA______AUUCAAAUUGCACCAUCAAAAAAUGUUUGUC_______________UACAUCUCGGCAAACACGGCGAG_____AAUUUC ................((((((.....................))))))..........((((((((.......................)))))))).................. ( -8.93 = -9.30 + 0.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:38:00 2011