| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,198,944 – 16,199,044 |
| Length | 100 |
| Max. P | 0.900621 |

| Location | 16,198,944 – 16,199,044 |
|---|---|
| Length | 100 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 67.35 |
| Shannon entropy | 0.69353 |
| G+C content | 0.49671 |
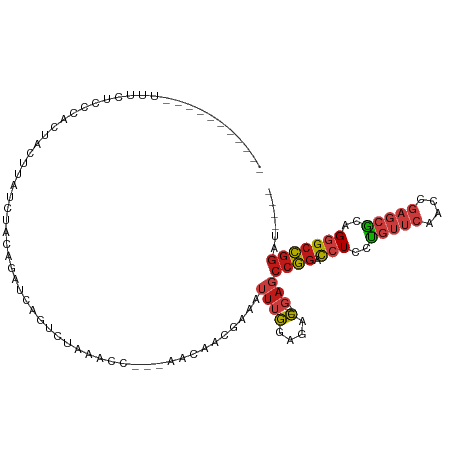
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -12.63 |
| Energy contribution | -13.17 |
| Covariance contribution | 0.54 |
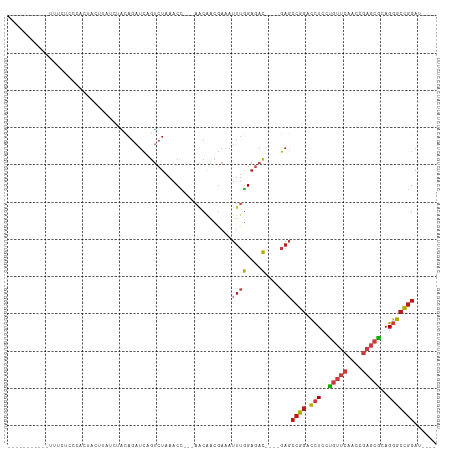
| Combinations/Pair | 1.33 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.900621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
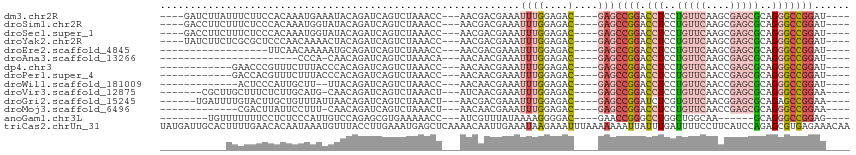
>dm3.chr2R 16198944 100 - 21146708 ----GAUCUUAUUUCUUCCACAAAUGAAAUACAGAUCAGUCUAAACC---AACGACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- ----(((((((((((..........)))))).))))).((((...((---((.........))))))))----...((((.(((..(((((....)))))..)))))))..---- ( -30.00, z-score = -2.61, R) >droSim1.chr2R 14857773 100 - 19596830 ----GACCUUCUUUCUCCCACAAAUGGUAUACAGAUCAGUCUAAACC---AACGACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- ----......((((((((......((((......))))(((......---...))).......))))).----)))((((.(((..(((((....)))))..)))))))..---- ( -25.50, z-score = -0.36, R) >droSec1.super_1 13764795 100 - 14215200 ----GACCUUCUUUCUCCCACAAAUGGUAUACAGAUCAGUCUAAACC---AACGACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- ----......((((((((......((((......))))(((......---...))).......))))).----)))((((.(((..(((((....)))))..)))))))..---- ( -25.50, z-score = -0.36, R) >droYak2.chr2R 8135180 100 + 21139217 ----UAUCUUCUCGCGCUCCCAACAAAACUACAGAUCAGUCUAAACC---AACGACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- ----.((((.(((((((((.(.......((.(((((..(((......---...)))...))))).))..----((((.((....)).))))..).)))))).)))..))))---- ( -27.30, z-score = -1.05, R) >droEre2.scaffold_4845 10350160 86 - 22589142 ------------------UUCAACAAAAAUGCAGAUCAGUCUAAACC---AACGACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- ------------------....................((((...((---((.........))))))))----...((((.(((..(((((....)))))..)))))))..---- ( -21.10, z-score = -0.43, R) >droAna3.scaffold_13266 16800339 80 + 19884421 -----------------------CCCA-CAACAGAUCAGUCUAAACA---AACAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAAGCGAGCGCAGGGCCGGAU---- -----------------------....-..........((((...((---((........)))).))))----...((((.(((..(((((....)))))..)))))))..---- ( -21.90, z-score = -1.56, R) >dp4.chr3 9856208 92 - 19779522 ------------GAACCCGUUUCUUUACCCACAGAUCAGUCUAAACC---AACAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAU---- ------------.....((((((.........(((....))).....---..(((......))))))))----)..((((.(((..(((((....)))))..)))))))..---- ( -22.60, z-score = -0.71, R) >droPer1.super_4 5173669 92 - 7162766 ------------GACCACGUUUCUUUACCCACAGAUCAGUCUAAACC---AACAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAU---- ------------.....((((((.........(((....))).....---..(((......))))))))----)..((((.(((..(((((....)))))..)))))))..---- ( -22.80, z-score = -0.89, R) >droWil1.scaffold_181009 1346698 89 - 3585778 -------------ACUCCCAUUGCUU--UUACAGAUCAGUCUAAACC---AACAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAU---- -------------.(((...(((...--...)))....((((...((---((.........))))))))----)))((((.(((..(((((....)))))..)))))))..---- ( -22.00, z-score = -0.72, R) >droVir3.scaffold_12875 9545640 96 + 20611582 -------CGCUUGCUUUCUCUUGCAUG-CAACAGAUCAGUCUAAACU---AUCAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAA---- -------.((.(((........))).)-)....(((.(((....)))---))).......((((....)----)))((((.(((..(((((....)))))..)))))))..---- ( -27.50, z-score = -0.82, R) >droGri2.scaffold_15245 13674742 98 - 18325388 ------UGAUUUUGUACUUGCUGUUUAUUAACAGAUCAGUCUAAACU---AACGACGAAAUUUGGAGAC----GAGCCGGAUCUCGUGUUCAACGGAGCGCAGAGCCGGAA---- ------..((((((((((..(((((....)))))...))).....(.---...))))))))(((....)----)).((((.(((.((((((....))))))))).))))..---- ( -28.10, z-score = -1.61, R) >droMoj3.scaffold_6496 15865015 90 - 26866924 -------------CGACUUAUUCCUUU-CAACAGAUCAGUCUAAACU---AACAACGAAAUUUGGAGAC----GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAA---- -------------.((((.(((.....-.....))).))))......---..........((((....)----)))((((.(((..(((((....)))))..)))))))..---- ( -22.10, z-score = -0.75, R) >anoGam1.chr3L 16272899 90 + 41284009 --------UGUUUUUUUCCUCUCCCAUUGUCCAGAGCGUGAAAAACC---AUCGUUUAUAAAAGGGGAC----GAACCGGGCCUGGCUGGCAA------GCAGGGCCGGAG---- --------.......(((.(((((..((((...(((((((......)---).)))))))))..))))).----)))((((.((((.((....)------))))).))))..---- ( -30.00, z-score = -0.81, R) >triCas2.chrUn_31 74669 115 + 298863 UAUGAUUGCACUUUUGAACACAAUAAAUGUUUACCUUGAAAUGAGCUCAAAACAAUUGAAAUAAGAAAUUUAAAAAAAUUAUUUGAUUUUCCUUCAUCCAGAGCGUGAGAAACAA ........(((...((((((.......))))))...........((((........((((....((((.(((((.......))))).)))).))))....)))))))........ ( -12.40, z-score = 0.74, R) >consensus ___________UUUCUCCCACUACUUAUCUACAGAUCAGUCUAAACC___AACAACGAAAUUUGGAGAC____GAGCCGGACCUCCUGUUCAACCGAGCGCAGGGCCGGAU____ ............................................................................((((.(((..(((((....)))))..)))))))...... (-12.63 = -13.17 + 0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:59 2011