| Sequence ID | dm3.chr2R |
|---|---|
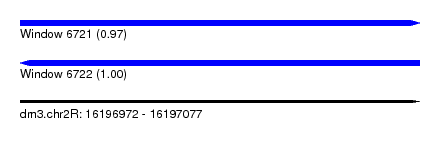
| Location | 16,196,972 – 16,197,077 |
| Length | 105 |
| Max. P | 0.999866 |

| Location | 16,196,972 – 16,197,077 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.67858 |
| G+C content | 0.48998 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
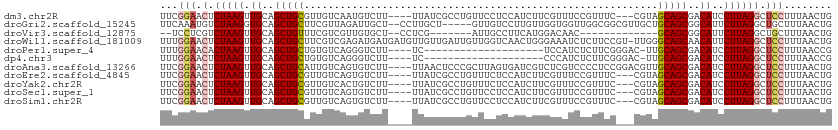
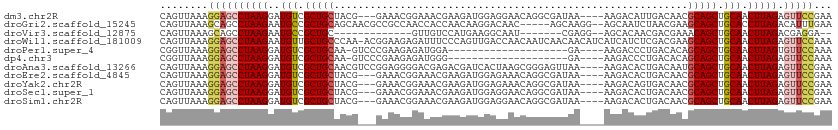
>dm3.chr2R 16196972 105 + 21146708 UUCGGAACUCUAAGUUGCAGCUGCGUUGUCAAUGUCUU----UUAUCGCCUGUUCCUCCAUCUUCGUUUCCGUUUC---CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((((((.((((((.((..((((....----......................))))..))....---)))))).)))))....))))).)))........ ( -19.27, z-score = -0.67, R) >droGri2.scaffold_15245 13672676 105 + 18325388 UUCAAAUGUCUAAGGUGCAGCUGCUUCGUUAGAUUGCU--CCUUGCU-----GUUGUCCUUGUUGGUGGUUGGCGGCGUUGCUGCAGCGGCAUUCUUAGGCUGCUUUAACUG .......(((((((((((.(((((.((....))..((.--...((((-----((((.((........)).))))))))..)).))))).))).))))))))........... ( -34.60, z-score = -0.96, R) >droVir3.scaffold_12875 9543649 88 - 20611582 --UCCUCGUCUAAGUUGCAGCUGUUUCGUCGUUGUGCU--CCUCG-------AUUGCCUUCAUGGACAAC-------------GCAGCGGCAUUCUUAGGCUGCUUUAACUG --..........((((((((((.....(((((((((.(--((..(-------(......))..)))...)-------------)))))))).......))))))...)))). ( -26.90, z-score = -2.01, R) >droWil1.scaffold_181009 1344694 111 + 3585778 UUUGGAACUCUAAGUUGCAGCUGCUUCGUCGAGAUGAUGAUGUUGUUGAUUGUUGGUCAACUGGGAAAUCUCUUCCGU-UUGGGCAGCAACAUUCUUAGGCUCCUUUAACUG ...(((.(.(((((.((((((.((.(((((.....))))).)).)))).((((((.((((.(((((......))))).-)))).))))))))..)))))).)))........ ( -32.90, z-score = -1.79, R) >droPer1.super_4 5171729 87 + 7162766 UUUGGAACACUAAGUUGCAGCUGCUGUGUCAGGGUCUU----UC--------------------UCCAUCUCUUCGGGAC-UUGCAGCGACAUCCUUAGGCUCCUUUAACCG ...(((.(.(((((.((..(((((...(((.(((....----..--------------------)))....(....))))-..)))))..))..)))))).)))........ ( -25.70, z-score = -1.58, R) >dp4.chr3 9854268 87 + 19779522 UUUGGAACUCUAAGUUGCAGCUGCUGUGUCAGGGUCUU----UC--------------------CCCAUCUCUUCGGGAC-UUGCAGCGACAUCCUUAGGCUCCUUUAACCG ...(((.(.(((((.((..(((((...(((.(((....----..--------------------)))....(....))))-..)))))..))..)))))).)))........ ( -28.20, z-score = -2.24, R) >droAna3.scaffold_13266 16798407 108 - 19884421 UUCGGAACUCUAAGUUGCAGCUGCAUUGUCAGUGUCUU----UUAACUCCCGCUUAGUGAUCGUCUCGUCCCCUCCGGACGUUGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((.((..((((((..((((.((....----............)).)))).....(((((.....))))).))))))..))..)))))).)))........ ( -28.29, z-score = -1.69, R) >droEre2.scaffold_4845 10348185 105 + 22589142 UUCGGAACUCUAAGUUGCAGCUGCGUUGUCAGUGUCUU----UUAUCGCCUGUUUCUCCAUCUUCGUUUCCGUUUC---CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((((((.((((((....(((.((...----.....)))))............((....))....---)))))).)))))....))))).)))........ ( -20.60, z-score = -0.80, R) >droYak2.chr2R 8133221 105 - 21139217 UUCGGAACUCUAAGUUGCAGCUGCGUUGUCACUGUCUU----UUAUCGCCUGUUUCUCCAUCUUCGUUUCCGUUUC---CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((.((..((((((..(..((.((...----.....))..))..)..)...........((....---))..)))))..))..)))))).)))........ ( -17.70, z-score = 0.04, R) >droSec1.super_1 13762826 105 + 14215200 UUCGGAACUCUAAGUUGCAGCUGCGUUGUCAGUGUCUU----UUAUCGCCUGUUCCUCCAUCUUCGUUUCCGUUUC---CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((((((.((((((....(((.((...----.....)))))............((....))....---)))))).)))))....))))).)))........ ( -20.60, z-score = -0.81, R) >droSim1.chr2R 14855807 105 + 19596830 UUCGGAACUCUAAGUUGCAGCUGCGUUGUCAGUGUCUU----UUAUCGCCUGUUCCUCCAUCUUCGUUUCCGUUUC---CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((((((.((((((....(((.((...----.....)))))............((....))....---)))))).)))))....))))).)))........ ( -20.60, z-score = -0.81, R) >consensus UUCGGAACUCUAAGUUGCAGCUGCGUUGUCAGUGUCUU____UUAUCGCCUGUUUCUCCAUCUUCGUUUCCGUUUC___CGUAGCAGCGACAUCCUUAGGCUCCUUUAACUG ...(((.(.(((((.((..(((((...........................................................)))))..))..)))))).)))........ (-13.18 = -13.50 + 0.32)
| Location | 16,196,972 – 16,197,077 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.91 |
| Shannon entropy | 0.67858 |
| G+C content | 0.48998 |
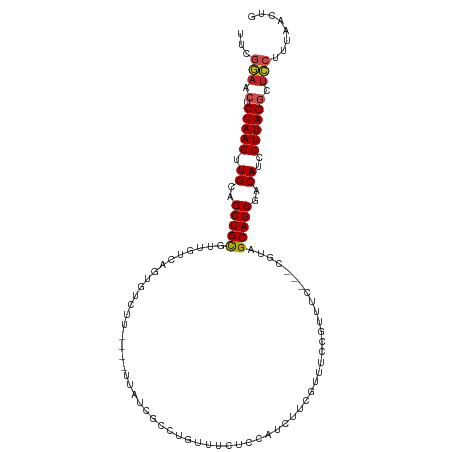
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.63 |
| SVM RNA-class probability | 0.999866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
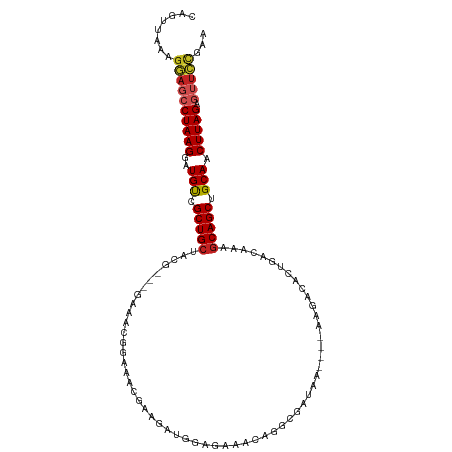
>dm3.chr2R 16196972 105 - 21146708 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAA----AAGACAUUGACAACGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.(((((..((---....))....((....((......))..))....----..............))))).))).))))).)))))... ( -31.80, z-score = -3.63, R) >droGri2.scaffold_15245 13672676 105 - 18325388 CAGUUAAAGCAGCCUAAGAAUGCCGCUGCAGCAACGCCGCCAACCACCAACAAGGACAAC-----AGCAAGG--AGCAAUCUAACGAAGCAGCUGCACCUUAGACAUUUGAA ...(((((...(.(((((..(((.(((((.((..(...((......((.....)).....-----.))..).--.))..((....)).))))).))).))))).).))))). ( -23.70, z-score = -0.72, R) >droVir3.scaffold_12875 9543649 88 + 20611582 CAGUUAAAGCAGCCUAAGAAUGCCGCUGC-------------GUUGUCCAUGAAGGCAAU-------CGAGG--AGCACAACGACGAAACAGCUGCAACUUAGACGAGGA-- ...........(.(((((..(((.((((.-------------.(((((..((...((...-------.....--.)).))..)))))..)))).))).))))).).....-- ( -20.30, z-score = 0.22, R) >droWil1.scaffold_181009 1344694 111 - 3585778 CAGUUAAAGGAGCCUAAGAAUGUUGCUGCCCAA-ACGGAAGAGAUUUCCCAGUUGACCAACAAUCAACAACAUCAUCAUCUCGACGAAGCAGCUGCAACUUAGAGUUCCAAA ........((((((((((..(((.(((((((..-..))..(((((......(((((.......))))).........)))))......))))).))).))))).)))))... ( -31.46, z-score = -3.79, R) >droPer1.super_4 5171729 87 - 7162766 CGGUUAAAGGAGCCUAAGGAUGUCGCUGCAA-GUCCCGAAGAGAUGGA--------------------GA----AAGACCCUGACACAGCAGCUGCAACUUAGUGUUCCAAA ........((((((((((..(((.(((((..-((((((......)))(--------------------(.----......)))))...))))).))).))))).)))))... ( -26.60, z-score = -1.91, R) >dp4.chr3 9854268 87 - 19779522 CGGUUAAAGGAGCCUAAGGAUGUCGCUGCAA-GUCCCGAAGAGAUGGG--------------------GA----AAGACCCUGACACAGCAGCUGCAACUUAGAGUUCCAAA ........((((((((((..(((.(((((..-(((.......)))(((--------------------..----....))).......))))).))).))))).)))))... ( -30.80, z-score = -2.74, R) >droAna3.scaffold_13266 16798407 108 + 19884421 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCAACGUCCGGAGGGGACGAGACGAUCACUAAGCGGGAGUUAA----AAGACACUGACAAUGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.((((((.(((((.....))))).............((((..(((..----..))).))).)..)))))).))).))))).)))))... ( -37.90, z-score = -3.32, R) >droEre2.scaffold_4845 10348185 105 - 22589142 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAA----AAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.(((((..((---....))....((....((......))..))....----..............))))).))).))))).)))))... ( -31.80, z-score = -3.71, R) >droYak2.chr2R 8133221 105 + 21139217 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAA----AAGACAGUGACAACGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..((((((((((.((---....))....((....((......))..))....----.)).))))))))..((....))..))))).)))))... ( -34.00, z-score = -4.34, R) >droSec1.super_1 13762826 105 - 14215200 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAA----AAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.(((((..((---....))....((....((......))..))....----..............))))).))).))))).)))))... ( -31.80, z-score = -3.47, R) >droSim1.chr2R 14855807 105 - 19596830 CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG---GAAACGGAAACGAAGAUGGAGGAACAGGCGAUAA----AAGACACUGACAACGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.(((((..((---....))....((....((......))..))....----..............))))).))).))))).)))))... ( -31.80, z-score = -3.47, R) >consensus CAGUUAAAGGAGCCUAAGGAUGUCGCUGCUACG___GAAACGGAAACGAAGAUGGAGAAACAGGCGAUAA____AAGACACUGACAAAGCAGCUGCAACUUAGAGUUCCGAA ........((((((((((..(((.(((((...........................................................))))).))).))))).)))))... (-18.90 = -19.25 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:58 2011