| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,120,289 – 16,120,383 |
| Length | 94 |
| Max. P | 0.994750 |

| Location | 16,120,289 – 16,120,383 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 76.70 |
| Shannon entropy | 0.52206 |
| G+C content | 0.59128 |
| Mean single sequence MFE | -36.39 |
| Consensus MFE | -33.39 |
| Energy contribution | -31.18 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.994750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
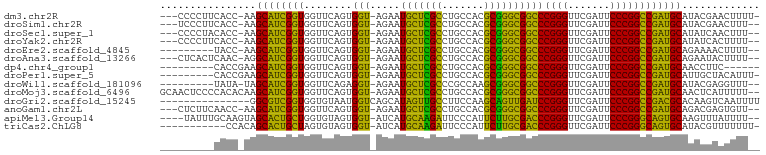
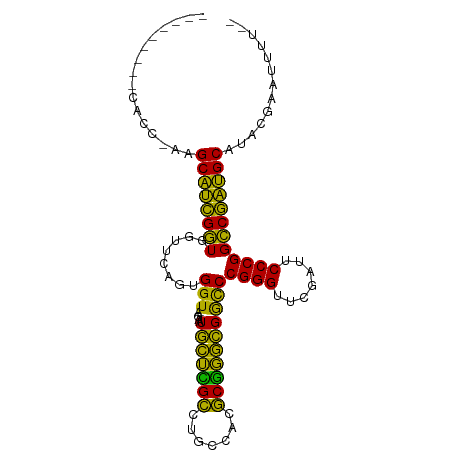
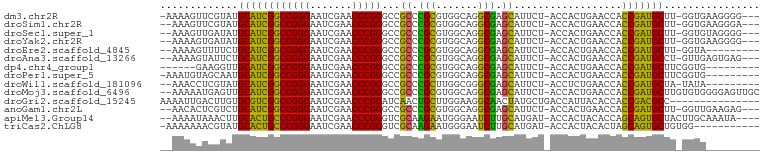
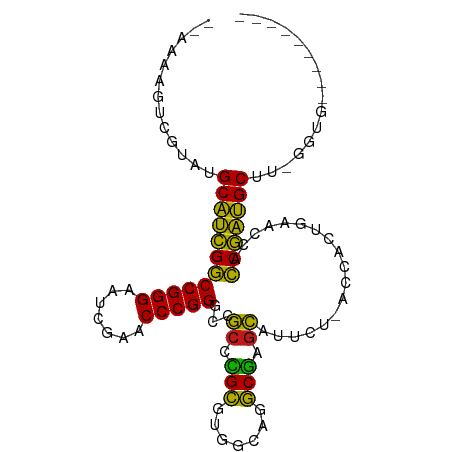
>dm3.chr2R 16120289 94 + 21146708 ---CCCCUUCACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAACUUUU- ---......((((-.......))))((((.(((((-((.........)))))))(((..(((((.(((.......)))))))).)))....))))....- ( -37.70, z-score = -1.48, R) >droSim1.chr2R 14778557 93 + 19596830 ---UCCCUUCACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAACUUU-- ---......((((-.......))))((((.(((((-((.........)))))))(((..(((((.(((.......)))))))).)))....))))...-- ( -37.70, z-score = -1.56, R) >droSec1.super_1 13686460 93 + 14215200 ---CCCCUACACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAUCAACUUU-- ---..........-..(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -35.90, z-score = -1.23, R) >droYak2.chr2R 8055703 93 - 21139217 ---CCCCUUCACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAUCACUUUU-- ---.....(((((-.......)))))...((((((-(...(((((((((((...))))))((((.(((.......))))))))).))))))))))...-- ( -37.60, z-score = -1.79, R) >droEre2.scaffold_4845 10270687 87 + 22589142 ---------UACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGAAAACUUUU-- ---------....-..(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -35.90, z-score = -1.61, R) >droAna3.scaffold_13266 3663054 93 - 19884421 ---CUCACUCAAC-AGGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGAAUACUUUU-- ---..........-..(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -35.70, z-score = -1.15, R) >dp4.chr4_group1 1123477 84 + 5278887 ---------CACCGAAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACCUUC------ ---------((((((....)))))).....(((((-((.........)))))))(((..(((((.(((.......)))))))).))).......------ ( -37.40, z-score = -1.71, R) >droPer1.super_5 2728467 89 - 6813705 ---------CACCGAAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUUGCUACAUUU- ---------((((((....)))))).....(((((-(..((((((((((((...))))))((((.(((.......))))))))).))))))))))....- ( -42.70, z-score = -3.00, R) >droWil1.scaffold_181096 7895065 87 - 12416693 ---------UAUA-UAGCAUCGGUGGUUCAGAGGU-AGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAGGUUU-- ---------....-..(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -38.10, z-score = -2.18, R) >droMoj3.scaffold_6496 10817791 97 - 26866924 GCAACUCCCCACACAAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACUCAUUUUU-- ................(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -35.90, z-score = -1.04, R) >droGri2.scaffold_15245 14888425 85 - 18325388 ---------------GGCGUCGGUGGUGUAAUGGUCAGCAUAGUUGCCUUCCAAGCAGUUGAUCCGGGUUCGAUUCCCGGCCGACGCACAAGUCAAUUUU ---------------.((((((((........(((((((...(((........))).)))))))((((.......))))))))))))............. ( -30.00, z-score = -2.17, R) >anoGam1.chr2L 33884288 93 - 48795086 ---CUCUUCAACC-AAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGACGAGUGUU-- ---..........-..(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...........-- ( -35.90, z-score = -0.55, R) >apiMel3.Group14 2180889 93 + 8318479 ----UAUUUGCAAGUAGCACUGCUGGUGUAGUGGU-AUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAAGUUUAUUUU-- ----.(((((((.(((.((((((....)))))).)-))...((((((.......))))))(.((((((.......)))))))..))))))).......-- ( -36.30, z-score = -4.11, R) >triCas2.ChLG8 12282825 87 + 15773733 -----------CCACAGCACUGCUAGUGUAGUGGU-AUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAUACGUUUUUUU- -----------......((((((....))))))((-(((((((((((.......))))))(.((((((.......))))))).))).))))........- ( -32.70, z-score = -3.92, R) >consensus _________CACC_AAGCAUCGGUGGUUCAGUGGU_AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAAUUUU__ ................((((((((........(((.....(((((((.......))))))))))((((.......))))))))))))............. (-33.39 = -31.18 + -2.21)
| Location | 16,120,289 – 16,120,383 |
|---|---|
| Length | 94 |
| Sequences | 14 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Shannon entropy | 0.52206 |
| G+C content | 0.59128 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -28.54 |
| Energy contribution | -27.01 |
| Covariance contribution | -1.53 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16120289 94 - 21146708 -AAAAGUUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUGAAGGGG--- -.....(((((..((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-.)))))....--- ( -33.70, z-score = -0.00, R) >droSim1.chr2R 14778557 93 - 19596830 --AAAGUUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUGAAGGGA--- --....(((((..((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-.)))))....--- ( -33.70, z-score = -0.03, R) >droSec1.super_1 13686460 93 - 14215200 --AAAGUUGAUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUGUAGGGG--- --...........((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-..........--- ( -32.20, z-score = 0.34, R) >droYak2.chr2R 8055703 93 + 21139217 --AAAAGUGAUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUGAAGGGG--- --...........((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-..........--- ( -32.20, z-score = 0.11, R) >droEre2.scaffold_4845 10270687 87 - 22589142 --AAAAGUUUUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUA--------- --...........((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-....--------- ( -32.20, z-score = -0.96, R) >droAna3.scaffold_13266 3663054 93 + 19884421 --AAAAGUAUUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCCU-GUUGAGUGAG--- --.....(((((.((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..-...)))))..--- ( -33.60, z-score = -1.24, R) >dp4.chr4_group1 1123477 84 - 5278887 ------GAAGGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUUCGGUG--------- ------...((((((.((((((.((........)).))))(((.((...)).))))))))....-))).......((((((....))))))--------- ( -33.20, z-score = -0.77, R) >droPer1.super_5 2728467 89 + 6813705 -AAAUGUAGCAAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUUCGGUG--------- -....((((.(((((.((((((.((........)).))))(((.((...)).))))))))))))-))........((((((....))))))--------- ( -37.40, z-score = -2.13, R) >droWil1.scaffold_181096 7895065 87 + 12416693 --AAACCUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCU-ACCUCUGAACCACCGAUGCUA-UAUA--------- --.......((((((((((((((((.......)))))(((((((((...))))))).)).....-............))))))).)-))).--------- ( -39.70, z-score = -3.85, R) >droMoj3.scaffold_6496 10817791 97 + 26866924 --AAAAAUGAGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUUGUGUGGGGAGUUGC --...........((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))................ ( -32.20, z-score = 0.66, R) >droGri2.scaffold_15245 14888425 85 + 18325388 AAAAUUGACUUGUGCGUCGGCCGGGAAUCGAACCCGGAUCAACUGCUUGGAAGGCAACUAUGCUGACCAUUACACCACCGACGCC--------------- .............((((((((((((.......)))))..........(((..((((....))))..)))........))))))).--------------- ( -28.90, z-score = -3.17, R) >anoGam1.chr2L 33884288 93 + 48795086 --AACACUCGUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUU-GGUUGAAGAG--- --..((..((...((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).)-)..)).....--- ( -34.00, z-score = -0.90, R) >apiMel3.Group14 2180889 93 - 8318479 --AAAAUAAACUUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAU-ACCACUACACCAGCAGUGCUACUUGCAAAUA---- --...........((((((((((((.......)))))...((((((.......)))))).....-............)))))))............---- ( -29.30, z-score = -3.15, R) >triCas2.ChLG8 12282825 87 - 15773733 -AAAAAAACGUAUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAU-ACCACUACACUAGCAGUGCUGUGG----------- -............((((((((((((.......)))))...((((((.......)))))).....-............))))))).....----------- ( -29.30, z-score = -2.73, R) >consensus __AAAAGUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU_ACCACUGAACCACCGAUGCUU_GGUG_________ .............((((((((((((.......)))))...((.(((.......))).))..................)))))))................ (-28.54 = -27.01 + -1.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:49 2011