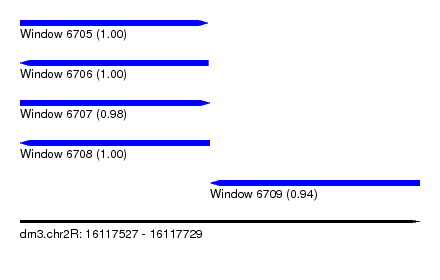
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,117,527 – 16,117,729 |
| Length | 202 |
| Max. P | 0.997710 |

| Location | 16,117,527 – 16,117,622 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.52823 |
| G+C content | 0.57479 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -17.93 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
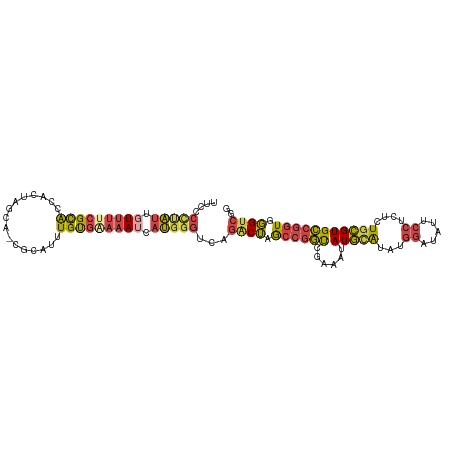
>dm3.chr2R 16117527 95 + 21146708 --------GCCAGAACCAAC---AAUAACAGGACUUGUU----GGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUUAAGCCGCC --------((..((((....---...(((((((...(((----((..(((....)))(((((((....)))))))..)))))....))))))).....))))..)).... ( -36.20, z-score = -3.74, R) >droSim1.chr2R 14775829 99 + 19596830 --------GCCAGAACCAAC---AAUAACAGGACUUGUUGGCCGGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUUAAGCCGCC --------((..((((....---...(((((((...((.(((.((..(((....)))(((((((....)))))))..)).))))).))))))).....))))..)).... ( -38.50, z-score = -3.31, R) >droSec1.super_1 13683702 99 + 14215200 --------GCCAGAACCAAC---AAUAACAGGACUUGUUGGCCGGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUUAAGCCGCC --------((..((((....---...(((((((...((.(((.((..(((....)))(((((((....)))))))..)).))))).))))))).....))))..)).... ( -38.50, z-score = -3.31, R) >droYak2.chr2R 8052795 96 - 21139217 --------GCCAGAACCAAC---AAUAACAGGACUUGUUU---GGCCGCCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUUAAGCCGCC --------((..((((....---...(((((((..((.((---((..((.....)).(((((((....)))))))..)))).))..))))))).....))))..)).... ( -30.50, z-score = -2.62, R) >droEre2.scaffold_4845 10267781 95 + 22589142 --------GCCAGAACCAAC---AAUAACAGGACUUGUU----GGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUCUAAGCCGCC --------((.(((......---...(((((((...(((----((..(((....)))(((((((....)))))))..)))))....)))))))......)))..)).... ( -34.76, z-score = -3.23, R) >droAna3.scaffold_13266 10658914 84 - 19884421 --------AGGAGGACUAGC---CGGGACAA-ACUAGGCG---AGCCGGCGGC--CAGGCUUGGCUAACCAAGCCAACCAGCCACUUCCUGUUUUAAAGCC--------- --------((((((....((---(((.....-........---..)))))(((--..(((((((....))))))).....))).))))))...........--------- ( -32.06, z-score = -1.29, R) >dp4.chr3 13254389 107 + 19779522 GCGCAACUGCCACAACUGGCGCUCCGAGCAGACAGUGCUAAGAGGUCGGAUGGC-UGGGCUUGGUUAGCCAAGCCAACCAGCCGUUUCCUGUUGCACAAUUUUAAGCU-- (.(((((........(((((.(((..((((.....))))..))))))))(((((-(((((((((....))))))...)))))))).....))))).)...........-- ( -43.90, z-score = -2.21, R) >droPer1.super_2 665709 107 - 9036312 GCGCAACUGCCACAACUGGCGCUCCGAGCAGACAGUGCUAAGAGGUCGGAUGGC-UGGGCUUGGUUAGCCAAGCCAACCAGCCGUUUCCUGUUGCACAAUUUUAAGCU-- (.(((((........(((((.(((..((((.....))))..))))))))(((((-(((((((((....))))))...)))))))).....))))).)...........-- ( -43.90, z-score = -2.21, R) >consensus ________GCCAGAACCAAC___AAUAACAGGACUUGUUG___GGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUUAAGCCGCC ..........................(((((.....(((....))).((((......(((((((....)))))))....)))).....)))))................. (-17.93 = -18.16 + 0.24)
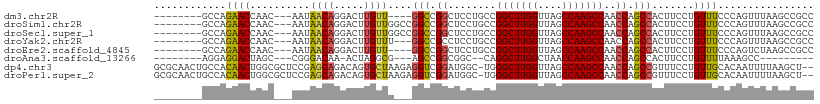
| Location | 16,117,527 – 16,117,622 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.74 |
| Shannon entropy | 0.52823 |
| G+C content | 0.57479 |
| Mean single sequence MFE | -43.55 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.95 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16117527 95 - 21146708 GGCGGCUUAAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCC----AACAAGUCCUGUUAUU---GUUGGUUCUGGC-------- (((.((((...(((.....))).)))).))).((((((((((....))))))))))....(((((((----(((((.........))---)))))..)))))-------- ( -42.50, z-score = -2.88, R) >droSim1.chr2R 14775829 99 - 19596830 GGCGGCUUAAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCCGGCCAACAAGUCCUGUUAUU---GUUGGUUCUGGC-------- ....(((..(((..(.((((((((..((((((((((((((((....)))))))(((....)))))))))))).....))))))..))---.)..)))..)))-------- ( -48.70, z-score = -3.63, R) >droSec1.super_1 13683702 99 - 14215200 GGCGGCUUAAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCCGGCCAACAAGUCCUGUUAUU---GUUGGUUCUGGC-------- ....(((..(((..(.((((((((..((((((((((((((((....)))))))(((....)))))))))))).....))))))..))---.)..)))..)))-------- ( -48.70, z-score = -3.63, R) >droYak2.chr2R 8052795 96 + 21139217 GGCGGCUUAAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGGCGGCC---AAACAAGUCCUGUUAUU---GUUGGUUCUGGC-------- ....(((..(((..(.((((((((..((((((((((((((((....))))))))))......)))))---)......))))))..))---.)..)))..)))-------- ( -39.60, z-score = -2.28, R) >droEre2.scaffold_4845 10267781 95 - 22589142 GGCGGCUUAGACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCC----AACAAGUCCUGUUAUU---GUUGGUUCUGGC-------- (((.((((...(((.....))).)))).))).((((((((((....))))))))))....(((((((----(((((.........))---)))))..)))))-------- ( -42.50, z-score = -2.68, R) >droAna3.scaffold_13266 10658914 84 + 19884421 ---------GGCUUUAAAACAGGAAGUGGCUGGUUGGCUUGGUUAGCCAAGCCUG--GCCGCCGGCU---CGCCUAGU-UUGUCCCG---GCUAGUCCUCCU-------- ---------...........((((.(.(((((((((((((((....))))))).)--)))(((((..---((......-.))..)))---)).)))))))))-------- ( -34.00, z-score = -0.92, R) >dp4.chr3 13254389 107 - 19779522 --AGCUUAAAAUUGUGCAACAGGAAACGGCUGGUUGGCUUGGCUAACCAAGCCCA-GCCAUCCGACCUCUUAGCACUGUCUGCUCGGAGCGCCAGUUGUGGCAGUUGCGC --...........(((((((......(((.((((((((((((....))))).)))-)))).)))..((((.((((.....)))).)))).((((....)))).))))))) ( -46.20, z-score = -2.71, R) >droPer1.super_2 665709 107 + 9036312 --AGCUUAAAAUUGUGCAACAGGAAACGGCUGGUUGGCUUGGCUAACCAAGCCCA-GCCAUCCGACCUCUUAGCACUGUCUGCUCGGAGCGCCAGUUGUGGCAGUUGCGC --...........(((((((......(((.((((((((((((....))))).)))-)))).)))..((((.((((.....)))).)))).((((....)))).))))))) ( -46.20, z-score = -2.71, R) >consensus GGCGGCUUAAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCC___CAACAAGUCCUGUUAUU___GUUGGUUCUGGC________ ...................(((((...(((((((((((((((....)))))))......))))))))....((((.....))))...........))))).......... (-23.32 = -23.95 + 0.63)

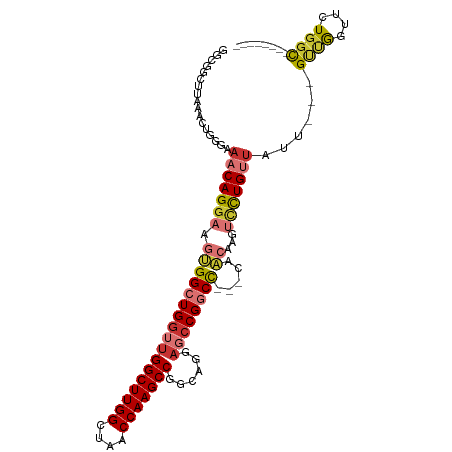
| Location | 16,117,527 – 16,117,623 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.35 |
| Shannon entropy | 0.61227 |
| G+C content | 0.57151 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -12.71 |
| Energy contribution | -12.58 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.977938 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
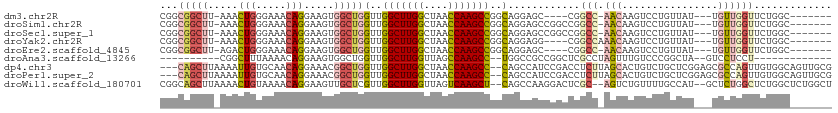
>dm3.chr2R 16117527 96 + 21146708 -------GCCAGAACCAACA---AUAACAGGACUUGUU-GGCCG----GCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUU-AAGCCGCCG -------((..((((.....---..(((((((...(((-((..(----((....)))(((((((....)))))))..)))))....))))))).....))))-..))..... ( -36.20, z-score = -3.76, R) >droSim1.chr2R 14775829 100 + 19596830 -------GCCAGAACCAACA---AUAACAGGACUUGUU-GGCCGGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUU-AAGCCGCCG -------((..((((.....---..(((((((...((.-(((.((..(((....)))(((((((....)))))))..)).))))).))))))).....))))-..))..... ( -38.50, z-score = -3.31, R) >droSec1.super_1 13683702 100 + 14215200 -------GCCAGAACCAACA---AUAACAGGACUUGUU-GGCCGGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUU-AAGCCGCCG -------((..((((.....---..(((((((...((.-(((.((..(((....)))(((((((....)))))))..)).))))).))))))).....))))-..))..... ( -38.50, z-score = -3.31, R) >droYak2.chr2R 8052795 97 - 21139217 -------GCCAGAACCAACA---AUAACAGGACUUGUUUGGCCG----CCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUU-AAGCCGCCG -------((..((((.....---..(((((((..((.((((..(----(.....)).(((((((....)))))))..)))).))..))))))).....))))-..))..... ( -30.50, z-score = -2.55, R) >droEre2.scaffold_4845 10267781 96 + 22589142 -------GCCAGAACCAACA---AUAACAGGACUUGUU-GGCCG----GCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUCU-AAGCCGCCG -------((.(((.......---..(((((((...(((-((..(----((....)))(((((((....)))))))..)))))....)))))))......)))-..))..... ( -34.76, z-score = -3.24, R) >droAna3.scaffold_13266 10658914 85 - 19884421 -------------AGGAGGAC--UAGCCGGGACAAACUAGGCGAGCCGGCGGCCA--GGCUUGGCUAACCAAGCCAACCAGCCACUUCCUGUUUUAAAGCCG---------- -------------((((((..--..(((((...............)))))(((..--(((((((....))))))).....))).))))))............---------- ( -32.06, z-score = -1.19, R) >dp4.chr3 13254390 107 + 19779522 CGCAACUGCCACAACUGGCGCUCCGAGCAGACAGUGCUAAGAGGUCGGAUGGCUG--GGCUUGGUUAGCCAAGCCAACCAGCCGUUUCCUGUUGCACAAUUUUAAGCUG--- .(((((........(((((.(((..((((.....))))..))))))))(((((((--(((((((....))))))...)))))))).....)))))..............--- ( -43.80, z-score = -2.40, R) >droPer1.super_2 665710 107 - 9036312 CGCAACUGCCACAACUGGCGCUCCGAGCAGACAGUGCUAAGAGGUCGGAUGGCUG--GGCUUGGUUAGCCAAGCCAACCAGCCGUUUCCUGUUGCACAAUUUUAAGCUG--- .(((((........(((((.(((..((((.....))))..))))))))(((((((--(((((((....))))))...)))))))).....)))))..............--- ( -43.80, z-score = -2.40, R) >droWil1.scaffold_180701 2619367 106 + 3904529 AGCCAGAGCCAGAGCCAGAGC--AUGGCAAAACAGACU--GCGAGUCCUUGGCUG--AGCUUGACUAACCAAGCCAACGAGCAACUUCCUGUUUUACAGUUUUAAGCUGCCG ((((((.((((..((....))--.))))......(((.--....))).))))))(--.(((((......))))))..((.(((.(((.(((.....)))....))).))))) ( -29.00, z-score = -0.41, R) >consensus _______GCCAGAACCAACA___AUAACAGGACUUGUU_GGCCGGCCGGCUCCUGCCGGCUUGGUUAGCCAAGCCAACCAGCCACUUCCUGUUUCCCAGUUU_AAGCCGCCG .........................(((((.........((...........))...(((((((....))))))).............)))))................... (-12.71 = -12.58 + -0.13)
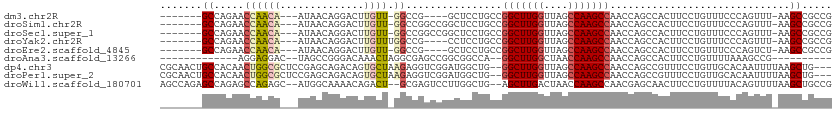
| Location | 16,117,527 – 16,117,623 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.35 |
| Shannon entropy | 0.61227 |
| G+C content | 0.57151 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -18.66 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.995596 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
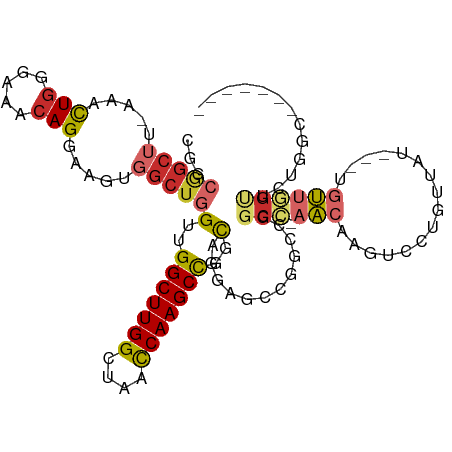
>dm3.chr2R 16117527 96 - 21146708 CGGCGGCUU-AAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGC----CGGCC-AACAAGUCCUGUUAU---UGUUGGUUCUGGC------- ((((.((((-...(((.....))).)))).))))((((((((((....))))))))))....((----(((((-(((((.........)---))))))..)))))------- ( -45.00, z-score = -3.54, R) >droSim1.chr2R 14775829 100 - 19596830 CGGCGGCUU-AAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCCGGCC-AACAAGUCCUGUUAU---UGUUGGUUCUGGC------- .....(((.-.(((..(.((((((((..((((((((((((((((....)))))))(((....)))))))))))-).....))))))..)---).)..)))..)))------- ( -48.70, z-score = -3.46, R) >droSec1.super_1 13683702 100 - 14215200 CGGCGGCUU-AAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCCGGCC-AACAAGUCCUGUUAU---UGUUGGUUCUGGC------- .....(((.-.(((..(.((((((((..((((((((((((((((....)))))))(((....)))))))))))-).....))))))..)---).)..)))..)))------- ( -48.70, z-score = -3.46, R) >droYak2.chr2R 8052795 97 + 21139217 CGGCGGCUU-AAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGG----CGGCCAAACAAGUCCUGUUAU---UGUUGGUUCUGGC------- .....(((.-.(((..(.((((((((..((((((((((((((((....))))))))))......----))))))......))))))..)---).)..)))..)))------- ( -39.60, z-score = -2.14, R) >droEre2.scaffold_4845 10267781 96 - 22589142 CGGCGGCUU-AGACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGC----CGGCC-AACAAGUCCUGUUAU---UGUUGGUUCUGGC------- ((((.((((-...(((.....))).)))).))))((((((((((....))))))))))....((----(((((-(((((.........)---))))))..)))))------- ( -45.00, z-score = -3.34, R) >droAna3.scaffold_13266 10658914 85 + 19884421 ----------CGGCUUUAAAACAGGAAGUGGCUGGUUGGCUUGGUUAGCCAAGCC--UGGCCGCCGGCUCGCCUAGUUUGUCCCGGCUA--GUCCUCCU------------- ----------............((((.(.(((((((((((((((....)))))))--.))))(((((..((.......))..))))).)--))))))))------------- ( -34.00, z-score = -0.85, R) >dp4.chr3 13254390 107 - 19779522 ---CAGCUUAAAAUUGUGCAACAGGAAACGGCUGGUUGGCUUGGCUAACCAAGCC--CAGCCAUCCGACCUCUUAGCACUGUCUGCUCGGAGCGCCAGUUGUGGCAGUUGCG ---..............(((((......(((.((((((((((((....))))).)--)))))).)))..((((.((((.....)))).)))).((((....)))).))))). ( -44.00, z-score = -2.33, R) >droPer1.super_2 665710 107 + 9036312 ---CAGCUUAAAAUUGUGCAACAGGAAACGGCUGGUUGGCUUGGCUAACCAAGCC--CAGCCAUCCGACCUCUUAGCACUGUCUGCUCGGAGCGCCAGUUGUGGCAGUUGCG ---..............(((((......(((.((((((((((((....))))).)--)))))).)))..((((.((((.....)))).)))).((((....)))).))))). ( -44.00, z-score = -2.33, R) >droWil1.scaffold_180701 2619367 106 - 3904529 CGGCAGCUUAAAACUGUAAAACAGGAAGUUGCUCGUUGGCUUGGUUAGUCAAGCU--CAGCCAAGGACUCGC--AGUCUGUUUUGCCAU--GCUCUGGCUCUGGCUCUGGCU (((((((((....(((.....))).))))))).))..(((((((....)))))))--.(((((.((((....--.)))).....((((.--((....))..))))..))))) ( -36.40, z-score = -0.82, R) >consensus CGGCGGCUU_AAACUGGGAAACAGGAAGUGGCUGGUUGGCUUGGCUAACCAAGCCGGCAGGAGCCGGCCGGCC_AACAAGUCCUGUUAU___UGUUGGUUCUGGC_______ ...(((((.....(((.....))).....)))))(..(((((((....)))))))..)...................................................... (-18.66 = -18.90 + 0.24)
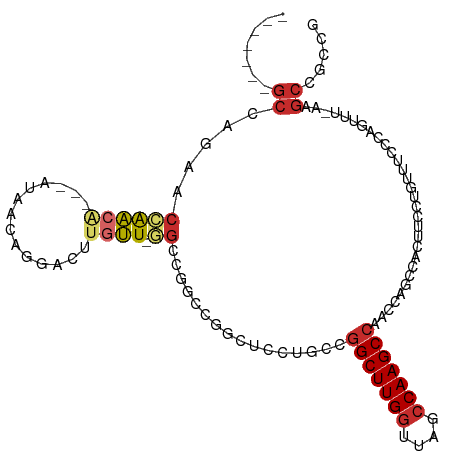
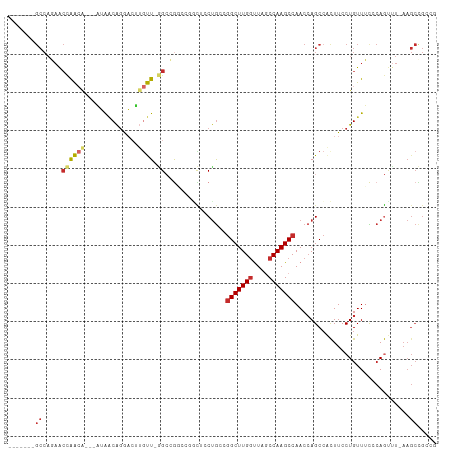
| Location | 16,117,623 – 16,117,729 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.52007 |
| G+C content | 0.49441 |
| Mean single sequence MFE | -32.96 |
| Consensus MFE | -22.41 |
| Energy contribution | -22.92 |
| Covariance contribution | 0.51 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
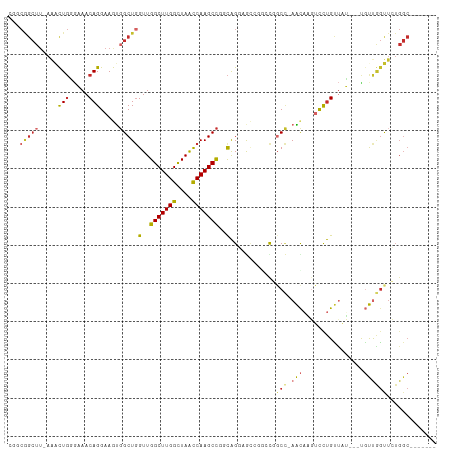
>dm3.chr2R 16117623 106 - 21146708 -UCCCCUAUU-UUUUCGCACCACUAGCA-CGCGUUUGUGAAAAUCAUGGGUGAGACUAGCCGGUCGAAAUAUGCAUAUGGAUAUUCCACUCUGCGUGCCGGUGGGUCGG -(((((....-((((((((..((.....-...)).))))))))....))).))((((.((((((......(((((..(((.....)))...))))))))))).)))).. ( -34.60, z-score = -1.50, R) >droSim1.chr2R 14775929 108 - 19596830 AUCCCCUAUUGUUUUCGCACCACUAGCA-CGCGUUUGUGCAAAUCAUGGGUCAGACUAGCCGGUGGAAAUAUGCAUAUGGAUAUUCCUCUCUGCGUGCCGGUGGGUCGG ...............((..((((..(((-(((...((((((..((((.(((.......))).)))).....)))))).((.....)).....))))))..))))..)). ( -35.50, z-score = -1.14, R) >droSec1.super_1 13683802 108 - 14215200 UUCCCCUAUUGUUUUCGCACCACUAGAA-CGCGUUUGCGAAAAUCAUGGGUCAGACUAGCCGGUGGAAAUAUGCAUAUGGAUAUUCCUCUCUGCGUGCCGGUGGGUCGG ....(((((.(((((((((..((.....-...)).))))))))).)))))...((((.((((((......(((((...((.....))....))))))))))).)))).. ( -36.30, z-score = -1.78, R) >droYak2.chr2R 8052892 109 + 21139217 UUUCCCCAUUGUUCUCGCACCUCUGGCACCGCAUUUGUGGAAAUCAUGGGUCAGGCUAGCCGGUCGAAAUAUGCAUAUGGAUAUUCCGCUCUACGUGCCGGUGGGUCGA ....(((((.(((..((((....((....))....))))..))).)))))...((((.((((((((.(((((.(....).))))).))........)))))).)))).. ( -31.40, z-score = 0.60, R) >droEre2.scaffold_4845 10267877 108 - 22589142 UGCCCUCAUUGUUUUCGCACCUCUAGCA-CGCAUUUGUGAAAAUCAUGGGUCAGGCUAGCCGGUCGAAAUAUGCAUAUGGAUAUACCUCUCUGCGUGCCGGUGGGUCGG ((.((.(((.(((((((((.........-......))))))))).))))).))((((.((((((......(((((...((.....))....))))))))))).)))).. ( -31.36, z-score = -0.00, R) >droWil1.scaffold_180701 2619473 103 - 3904529 ----UUUGCU-UUUUCGUGUGCAUAUUU-UGAAUCUGCACAUACACCUUCCCCCACAUAGUCACACUAAUAUAUGUUCACAUAUGUAUUAAUGUGUGUUUGUGUGCCGG ----......-.....(((((((.((..-...)).))))))).......((..(((((((.(((((((((((((((....))))))))))..))))).)))))))..)) ( -28.60, z-score = -3.65, R) >consensus UUCCCCUAUUGUUUUCGCACCACUAGCA_CGCAUUUGUGAAAAUCAUGGGUCAGACUAGCCGGUCGAAAUAUGCAUAUGGAUAUUCCUCUCUGCGUGCCGGUGGGUCGG ....(((((.(((((((((................))))))))).)))))...((((.((((((......(((((...((.....))....))))))))))).)))).. (-22.41 = -22.92 + 0.51)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:47 2011