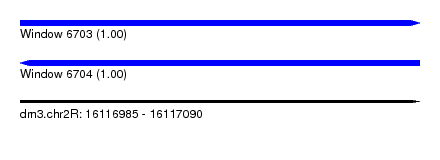
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,116,985 – 16,117,090 |
| Length | 105 |
| Max. P | 0.998591 |

| Location | 16,116,985 – 16,117,090 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.25 |
| Shannon entropy | 0.70228 |
| G+C content | 0.51192 |
| Mean single sequence MFE | -36.14 |
| Consensus MFE | -29.06 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
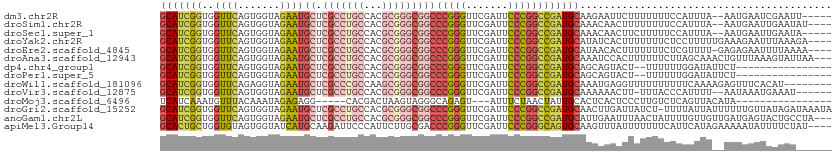
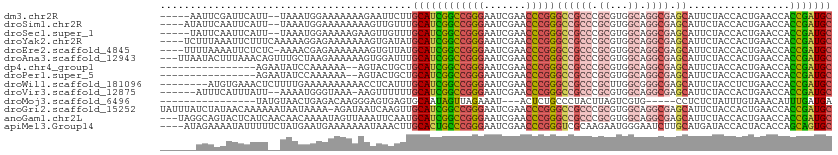
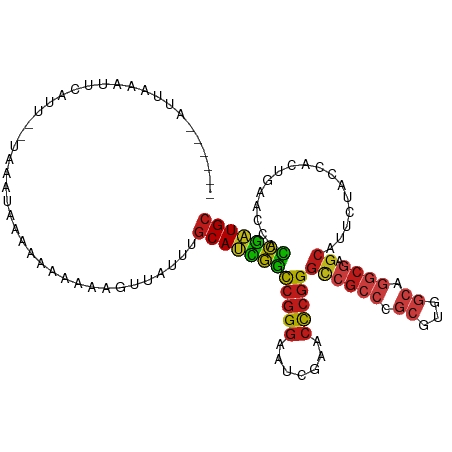
>dm3.chr2R 16116985 105 + 21146708 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAGAAUUCUUUUUUUCCAUUUA--AAUGAAUCGAAUU----- (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............(((.((((.--...)))).)))..----- ( -36.40, z-score = -1.05, R) >droSim1.chr2R 14775291 106 + 19596830 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACAACUUUUUUUUCCAUUUA--AAUGAAUUGAAUAU---- (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............(((.((((.--...)))).)))...---- ( -36.40, z-score = -1.43, R) >droSec1.super_1 13681791 105 + 14215200 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACAACUUCUUUUUCCAUUUA--AAUGAAUUGAAUA----- (((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...(((.(((.(((.......)--)).))))))....----- ( -36.50, z-score = -1.40, R) >droYak2.chr2R 8052230 108 - 21139217 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAUCACUUUUUUUCUCCUUUUUGAAAGAAUUUAAAGA---- (((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))........(((((((........)))))))........---- ( -39.40, z-score = -2.08, R) >droEre2.scaffold_4845 10267225 107 + 22589142 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUAACACUUUUUUUCUCGUUUU-GAGAGAAUUUUAAAA---- (((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))..........(((((((.....-)))))))........---- ( -41.20, z-score = -2.53, R) >droAna3.scaffold_12943 1998866 109 - 5039921 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUCCACUUUUUUCUUAGCAAACUGUUUAAAGUAUUAA--- (((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))......(((((.....(((....)))...))))).....--- ( -37.60, z-score = -1.55, R) >dp4.chr4_group1 1122842 94 + 5278887 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGCAGUACU--UUUUUUGGAUAUUCU---------------- (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).........--...............---------------- ( -35.70, z-score = -0.88, R) >droPer1.super_5 2727832 94 - 6813705 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGCAGUACU--UUUUUUGGAUAUUCU---------------- (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).........--...............---------------- ( -35.70, z-score = -0.88, R) >droWil1.scaffold_181096 7890325 104 - 12416693 GCAUCGGUGGUUCAGAGGUAGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUGAGGUUUUUUUUUUCAAAAGAGUUUCACAU-------- (((((((..((((.......))))((.(((((((...)))))))))(((((.......))))))))))))...(((..(((((((....)))))))..)))...-------- ( -43.50, z-score = -3.26, R) >droVir3.scaffold_12875 10537209 103 + 20611582 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAAACUU-UUUACCCAUUUU--AAUAAAUGAAAU------ (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).........-......(((((.--...)))))....------ ( -37.30, z-score = -1.95, R) >droMoj3.scaffold_6496 21774806 88 + 26866924 UCAUCAAAUGUUUACAAAUAGAGAGG-----CACGACUAAGUAGGGCAGAGU---AUUUCUAACUAUUGCACUCACUCCCUUGUCUCAGUUACAUA---------------- ......................((((-----((.(....(((..((((((((---.......))).)))).)..)))..).)))))).........---------------- ( -14.70, z-score = 0.10, R) >droGri2.scaffold_15252 9035298 111 - 17193109 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACUUGAUUAUCU-UUUUAUUAUUUUUUGUUAUAGAUAAAUA (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).......((((((-....((........))...))))))... ( -38.00, z-score = -2.04, R) >anoGam1.chr2L 33881892 109 - 48795086 GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUUGAAUUUAACUAUUUUGUUGUUGAUGAGUACUGCCUA--- .....((..(((((.......(((((((((((((...))))))((((.(((.......))))))))).)))))....(((((.........))))))))..))..))..--- ( -38.30, z-score = -0.94, R) >apiMel3.Group14 2180901 108 + 8318479 GCACUGCUGGUGUAGUGGUAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAAGUUUAUUUUUUUCAUUCAUAGAAAAAUAUUUUCUAU---- ((((((((((((......)))))(((((((.......)))))))..(((((.......))))))))))))...................(((((((.....)))))))---- ( -35.20, z-score = -3.95, R) >consensus GCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAAAAUUUUUUUUUCAUUUA__AAUGAAUUGAAU______ (((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).......................................... (-29.06 = -29.90 + 0.84)
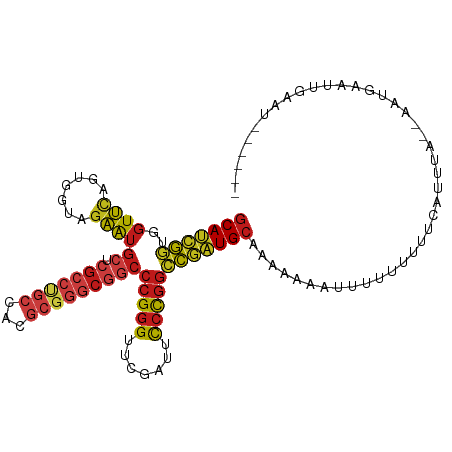
| Location | 16,116,985 – 16,117,090 |
|---|---|
| Length | 105 |
| Sequences | 14 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.25 |
| Shannon entropy | 0.70228 |
| G+C content | 0.51192 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -25.71 |
| Energy contribution | -26.16 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.997643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2R 16116985 105 - 21146708 -----AAUUCGAUUCAUU--UAAAUGGAAAAAAAGAAUUCUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC -----..(((.(((....--..))).))).............((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -32.70, z-score = -1.08, R) >droSim1.chr2R 14775291 106 - 19596830 ----AUAUUCAAUUCAUU--UAAAUGGAAAAAAAAGUUGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ----........(((((.--...)))))..............((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -32.30, z-score = -0.96, R) >droSec1.super_1 13681791 105 - 14215200 -----UAUUCAAUUCAUU--UAAAUGGAAAAAGAAGUUGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC -----.......(((((.--...)))))..............((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -32.30, z-score = -0.83, R) >droYak2.chr2R 8052230 108 + 21139217 ----UCUUUAAAUUCUUUCAAAAAGGAGAAAAAAAGUGAUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ----.((((...((((((......))))))..))))......((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -33.90, z-score = -1.50, R) >droEre2.scaffold_4845 10267225 107 - 22589142 ----UUUUAAAAUUCUCUC-AAAACGAGAAAAAAAGUGUUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ----........(((((..-.....)))))............((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -34.90, z-score = -2.07, R) >droAna3.scaffold_12943 1998866 109 + 5039921 ---UUAAUACUUUAAACAGUUUGCUAAGAAAAAAGUGGAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ---..............((((..((........))..)))).((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -35.10, z-score = -1.60, R) >dp4.chr4_group1 1122842 94 - 5278887 ----------------AGAAUAUCCAAAAAA--AGUACUGCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ----------------...............--.........((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -31.00, z-score = -1.16, R) >droPer1.super_5 2727832 94 + 6813705 ----------------AGAAUAUCCAAAAAA--AGUACUGCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ----------------...............--.........((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -31.00, z-score = -1.16, R) >droWil1.scaffold_181096 7890325 104 + 12416693 --------AUGUGAAACUCUUUUGAAAAAAAAAACCUCAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCUACCUCUGAACCACCGAUGC --------..((((.....((((....)))).....))))..((((((((((((.......)))))(((((((((...))))))).)).................))))))) ( -38.90, z-score = -3.46, R) >droVir3.scaffold_12875 10537209 103 - 20611582 ------AUUUCAUUUAUU--AAAAUGGGUAAA-AAGUUUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ------..(((((((...--.)))))))....-.........((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -33.20, z-score = -1.12, R) >droMoj3.scaffold_6496 21774806 88 - 26866924 ----------------UAUGUAACUGAGACAAGGGAGUGAGUGCAAUAGUUAGAAAU---ACUCUGCCCUACUUAGUCGUG-----CCUCUCUAUUUGUAAACAUUUGAUGA ----------------...(..((((((...((((...(((((.............)---))))..)))).))))))..).-----.......................... ( -15.02, z-score = 0.76, R) >droGri2.scaffold_15252 9035298 111 + 17193109 UAUUUAUCUAUAACAAAAAAUAAUAAAA-AGAUAAUCAAGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ...((((((...................-)))))).......((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -32.41, z-score = -2.16, R) >anoGam1.chr2L 33881892 109 + 48795086 ---UAGGCAGUACUCAUCAACAACAAAAUAGUUAAAUUCAAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ---.......................................((((((((((((.......)))))((((((.((...)).)))).)).................))))))) ( -31.00, z-score = -0.76, R) >apiMel3.Group14 2180901 108 - 8318479 ----AUAGAAAAUAUUUUUCUAUGAAUGAAAAAAAUAAACUUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUACCACUACACCAGCAGUGC ----(((((((.....)))))))...................((((((((((((.......)))))...((((((.......)))))).................))))))) ( -31.70, z-score = -3.71, R) >consensus ______AUUAAAUUCAUU__UAAAUAAAAAAAAAAGUUAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGC ..........................................((((((((((((.......)))))((((((.((...)).)))).)).................))))))) (-25.71 = -26.16 + 0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:43 2011