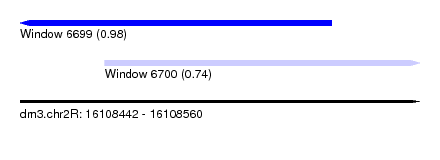
| Sequence ID | dm3.chr2R |
|---|---|
| Location | 16,108,442 – 16,108,560 |
| Length | 118 |
| Max. P | 0.979326 |

| Location | 16,108,442 – 16,108,534 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 73.49 |
| Shannon entropy | 0.55273 |
| G+C content | 0.43471 |
| Mean single sequence MFE | -13.98 |
| Consensus MFE | -10.54 |
| Energy contribution | -10.54 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
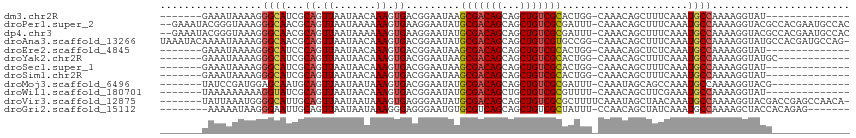
>dm3.chr2R 16108442 92 - 21146708 -UGCCAGUGCGACAGCUGCUGUCGCUUAUUCCGUCACUUUGUUAUUAACUGCGAUGCCCUUUUAUUUCUUUCAUUUCUUGCAUUUCACCCCAC -(((.((.(((((((...)))))))......((((.(...((.....)).).))))....................)).)))........... ( -14.70, z-score = -1.32, R) >droPer1.super_2 657480 87 + 9036312 -UGAAAUCGCGACAGCUGCUGUCGCAUAUUCCUUCACUUUUUUAUUAACUGCGUUGCCCU----UUACCCGUAUUUCUUUUAUUCUCAACCG- -.((((..(((((((...)))))))........................((((.......----.....))))))))...............- ( -12.50, z-score = -2.03, R) >dp4.chr3 13246116 87 - 19779522 -UGAAAUCGCGACAGCUGCUGUCGCAUAUUCCUUCACUUUUUUAUUAACUGCGUUGCCCU----UUACCCGUAUUUCUUUUAUUCUCAACCG- -.((((..(((((((...)))))))........................((((.......----.....))))))))...............- ( -12.50, z-score = -2.03, R) >droYak2.chr2R 8043618 90 + 21139217 -UGCCAGUGCGACAGCUGCUGUCGCUUAUUCCGUCACUUUGUUAUUAACUGCGAUGCCCUUUUAUUUCUUUCACUUCUUGCAUUUCAACCC-- -(((.((.(((((((...)))))))......((((.(...((.....)).).))))....................)).))).........-- ( -14.70, z-score = -1.22, R) >droSec1.super_1 13668745 91 - 14215200 -UGCCAGUGCGACAGCUGCUGUCGCUUAUUCCGUCACUUUGUUAUUAACUGCGAUGCCCUUUUAUUUCUUUCAUUCCUUGCAUUUCACCCCC- -(((.((.(((((((...)))))))......((((.(...((.....)).).))))....................)).)))..........- ( -14.90, z-score = -1.78, R) >droSim1.chr2R 14766917 91 - 19596830 -UGCCAGUGCGACAGCUGCUGUCGCUUAUUCCGUCACUUUGUUAUUAACUGCGAUGCCCUUUUAUUUCUUUCAUUCCUUGCAUUUCACCCCC- -(((.((.(((((((...)))))))......((((.(...((.....)).).))))....................)).)))..........- ( -14.90, z-score = -1.78, R) >droMoj3.scaffold_6496 21761324 92 - 26866924 -UGAAAUCGCGACAGCUGCUGUCGCAUAUUCCGUCACUUUAUUAUUAACUGCAUUGCUCCAUCGGAUACUCUAACAAGUGCUUUCCAUUCGAC -.......(((((((...))))))).......(((...............((...))......(((((((......))))...)))....))) ( -15.70, z-score = -0.28, R) >droWil1.scaffold_180701 1304113 84 + 3904529 -UGAAAACGCGACAGCAGCUGUCGCAUAUUCCGUCACUUUGUUAUUAACUGCGAUACCUUUUUUUUUACCUCAGACAGUGAAUUG-------- -.......(((((((...)))))))........(((((((((........))))......................)))))....-------- ( -13.84, z-score = -0.31, R) >droVir3.scaffold_12875 17233333 81 + 20611582 UGAAAAGCGCGACAGCUGCUGUCGCAUAUUCCCUCACUUUAUUAUUAACUGCAAUGCCCCAUUUAAUACCCUAACAUCUGG------------ ........(((((((...))))))).................((((((.((........)).)))))).............------------ ( -12.10, z-score = -0.91, R) >consensus _UGAAAGCGCGACAGCUGCUGUCGCAUAUUCCGUCACUUUGUUAUUAACUGCGAUGCCCUUUUAUUUCCUUCAUUUCUUGCAUUUCAACCC__ ........(((((((...))))))).................................................................... (-10.54 = -10.54 + 0.00)
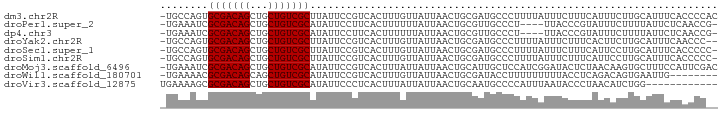
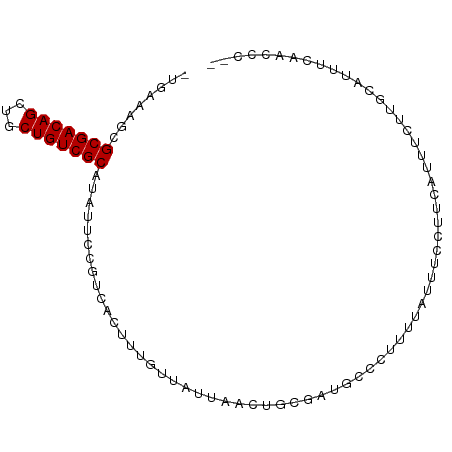
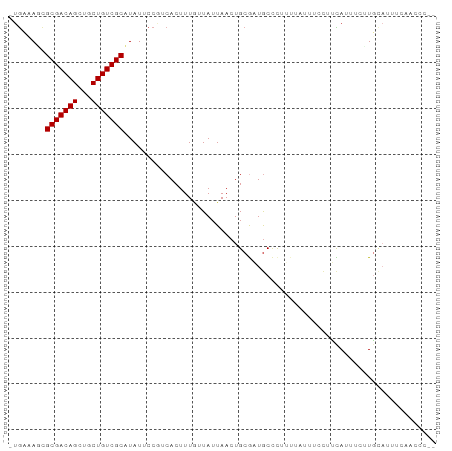
| Location | 16,108,467 – 16,108,560 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 77.25 |
| Shannon entropy | 0.46540 |
| G+C content | 0.43055 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.735367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
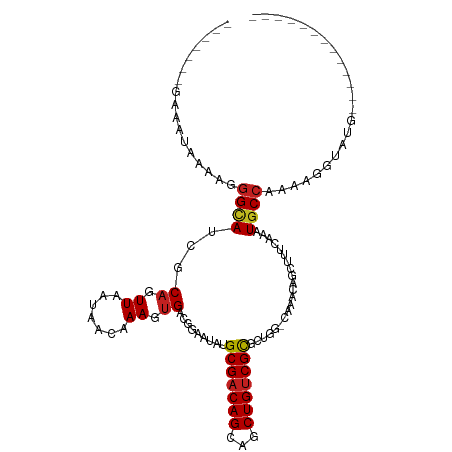
>dm3.chr2R 16108467 93 + 21146708 -------GAAAUAAAAGGGCAUCGCAGUUAAUAACAAAGUGACGGAAUAAGCGACAGCAGCUGUCGCACUGG-CAAACAGCUUUCAAAUGCCAAAAGGUAU-------------- -------..........(((((((((.((.......)).)).))(((...(((((((...))))))).(((.-....)))..)))..))))).........-------------- ( -21.70, z-score = -1.42, R) >droPer1.super_2 657495 112 - 9036312 --GAAAUACGGGUAAAGGGCAACGCAGUUAAUAAAAAAGUGAAGGAAUAUGCGACAGCAGCUGUCGCGAUUU-CAAACAGCUUUCAAAUGCCAAAAGGUACGCCACGAAUGCCAC --........((((..((((...............(((((...(((((.((((((((...))))))))))))-).....)))))....((((....)))).))).)...)))).. ( -29.20, z-score = -1.91, R) >dp4.chr3 13246131 112 + 19779522 --GAAAUACGGGUAAAGGGCAACGCAGUUAAUAAAAAAGUGAAGGAAUAUGCGACAGCAGCUGUCGCGAUUU-CAAACAGCUUUCAAAUGCCAAAAGGUACGCCACGAAUGCCAC --........((((..((((...............(((((...(((((.((((((((...))))))))))))-).....)))))....((((....)))).))).)...)))).. ( -29.20, z-score = -1.91, R) >droAna3.scaffold_13266 10650449 113 - 19884421 UAAAUACAAAAUAAAAGGGCAACGCAGUUAAUAACAAAGUGACGGAAUAUGCGACAGCAGCUGUCGUGCCGG-CAAACAGCUUUCAAAUGCCAAAAGGUAUGCCACGAUGCCAG- .................((((.((..((............(((((....(((....))).)))))(((((((-((.............))))....)))))))..)).))))..- ( -26.02, z-score = -0.42, R) >droEre2.scaffold_4845 10258645 93 + 22589142 -------GAAAUAAAAGGGCAUCCCAGUUAAUAACAAAGUGACGGAAUAAGCGACAGCAGCUGUCGCACUGG-CAAACAGCUCUCAAAUGCCAAAAGGUAU-------------- -------........(((((.(((..((((.........)))))))....(((((((...))))))).....-......)))))...(((((....)))))-------------- ( -22.60, z-score = -1.72, R) >droYak2.chr2R 8043641 95 - 21139217 -------GAAAUAAAAGGGCAUCGCAGUUAAUAACAAAGUGACGGAAUAAGCGACAGCAGCUGUCGCACUGG-CAAACAGCUUUCAAAUGCCAAAAGGUAUGC------------ -------..........(((((((((.((.......)).)).))(((...(((((((...))))))).(((.-....)))..)))..)))))...........------------ ( -21.70, z-score = -0.75, R) >droSec1.super_1 13668769 93 + 14215200 -------GAAAUAAAAGGGCAUCGCAGUUAAUAACAAAGUGACGGAAUAAGCGACAGCAGCUGUCGCACUGG-CAAACAGCUUUCAAAUGCCAAAAGGUAU-------------- -------..........(((((((((.((.......)).)).))(((...(((((((...))))))).(((.-....)))..)))..))))).........-------------- ( -21.70, z-score = -1.42, R) >droSim1.chr2R 14766941 93 + 19596830 -------GAAAUAAAAGGGCAUCGCAGUUAAUAACAAAGUGACGGAAUAAGCGACAGCAGCUGUCGCACUGG-CAAACAGCUUUCAAAUGCCAAAAGGUAU-------------- -------..........(((((((((.((.......)).)).))(((...(((((((...))))))).(((.-....)))..)))..))))).........-------------- ( -21.70, z-score = -1.42, R) >droMoj3.scaffold_6496 21761349 94 + 26866924 -------UAUCCGAUGGAGCAAUGCAGUUAAUAAUAAAGUGACGGAAUAUGCGACAGCAGCUGUCGCGAUUU-CAAAUAGCAGCCAAAUGCCAAAAGGUACG------------- -------.......(((.....(((.((((.........))))(((((.((((((((...))))))))))))-).....))).)))..((((....))))..------------- ( -24.20, z-score = -1.71, R) >droWil1.scaffold_180701 1304130 93 - 3904529 -------UAAAAAAAAAGGUAUCGCAGUUAAUAACAAAGUGACGGAAUAUGCGACAGCUGCUGUCGCGUUUU-CAAACAGCUUCGAAAUGCCAAAAGGUAU-------------- -------..........(((((..(.((.....)).((((....(((.(((((((((...))))))))).))-).....)))).)..))))).........-------------- ( -23.20, z-score = -2.18, R) >droVir3.scaffold_12875 17233346 107 - 20611582 -------UAUUAAAUGGGGCAUUGCAGUUAAUAAUAAAGUGAGGGAAUAUGCGACAGCAGCUGUCGCGCUUUUCAAAUAGCUAACAAAUGCCAAAAGGUACGACCGAGCCAACA- -------.......(((((((((...((((........((((.((....(((....))).)).))))(((........))))))).))))))....((.....))...)))...- ( -28.00, z-score = -1.51, R) >droGri2.scaffold_15112 2769968 99 + 5172618 --------AAAAAUAAGGGAAUUGCAGUUAAUAAUAAAGGGAGGGAAUGUGCGUCAGCAGCUGUCGCUAUUU-CCAACAGCUAUCAAAUGCCAAAAGCUACCACAGAG------- --------........((.....((..............(..(((((((.(((.(((...))).))))))))-))..).((........)).....))..))......------- ( -19.20, z-score = 0.26, R) >consensus _______GAAAUAAAAGGGCAUCGCAGUUAAUAACAAAGUGACGGAAUAUGCGACAGCAGCUGUCGCGCUGG_CAAACAGCUUUCAAAUGCCAAAAGGUAUG_____________ .................((((...((.((.......)).)).........(((((((...))))))).....................))))....................... (-14.15 = -14.33 + 0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:37:40 2011