| Sequence ID | dm3.chr2L |
|---|---|
| Location | 3,393,029 – 3,393,127 |
| Length | 98 |
| Max. P | 0.500000 |

| Location | 3,393,029 – 3,393,127 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 68.80 |
| Shannon entropy | 0.63180 |
| G+C content | 0.40145 |
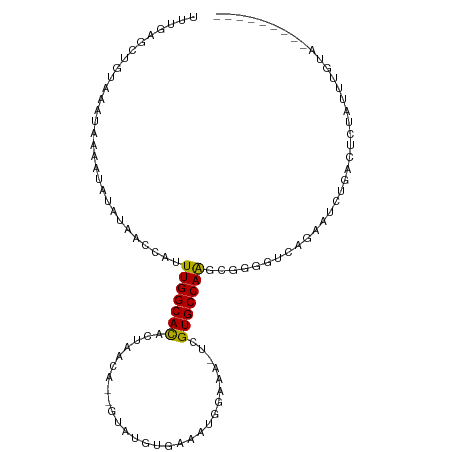
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -8.17 |
| Energy contribution | -7.90 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
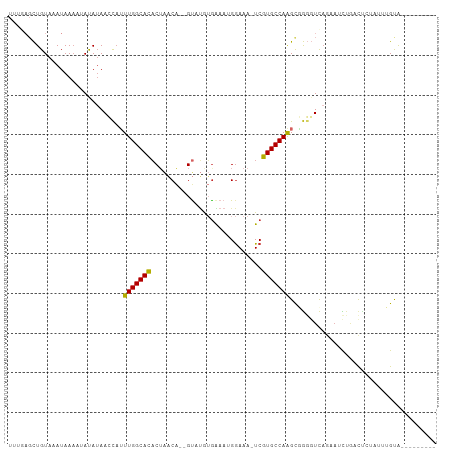
>dm3.chr2L 3393029 98 + 23011544 UUUGAGCUGUAAAUAAAAUAUAUAACCAUUUGGCACACUAACA--GUAUGUGAAAUGGAAA-UCGUGCCAAGCGGGGUCAGAAUCUGACUCUCUAUGUAUG------- .................(((((((.((((((.(((.((.....--)).))).))))))...-.(((.....)))(((((((...)))))))..))))))).------- ( -24.00, z-score = -1.92, R) >droWil1.scaffold_180708 1659538 106 - 12563649 UGAAAGUUGUAAAUAAA--ACAUAAGCUUUUGGCACUCUGUCGAUGCGAAGGAAAGGAAAAAUCGUGCCAGGCAACUUUCCAAGUCCUCUGAGUUGGCACUGUCAACA .(((((((((.......--..........((((((((((.(((...))).)))...((....))))))))))))))))))............((((((...)))))). ( -25.42, z-score = -0.47, R) >droPer1.super_1 2623186 96 - 10282868 -----UCUGUAAAUAAA--ACAUAAGCAGUUGGCAUACAAACACGGUAUAUGGAAAGGAAA-UCGUGCCAAGCCGGGUCAACAGUCAUUC----UGGCACUGUCAACA -----............--.........(((((((......((((((...(....)....)-)))))....((((((...........))----))))..))))))). ( -21.70, z-score = -1.04, R) >dp4.chr4_group3 5525481 96 - 11692001 -----UCUGUAAAUAAA--ACAUAAGCAGUUGGCAUACAAACACGGUAUAUGGAAAGGAAA-UCGUGCCAAGCCGGGUCAACAGUCAUUC----UGGCACUGUCAACA -----............--.........(((((((......((((((...(....)....)-)))))....((((((...........))----))))..))))))). ( -21.70, z-score = -1.04, R) >droSec1.super_5 1539822 96 + 5866729 UUUGAGCUGUAAAUAAAAUAUAUAACCAUUUGGCACACUAACA--GUAUGUGAAAUGGAAA-UCGUGCCAAGCGGGGUCAGAAUCUGACUCUAUUUGCG--------- ........(((((((..........((((((.(((.((.....--)).))).))))))...-.(((.....)))(((((((...)))))))))))))).--------- ( -25.70, z-score = -2.58, R) >droYak2.chr2L 3378137 104 + 22324452 UUUGAGCUGUAAAUAAUAUAUAUAACCAUUUGGCACACUAACA--GUAUGUGAAAUGGAAA-UCGUGCCAAGCGGUUUCAAAAUCUGAUCCUAUAUGUAUGUGUAUG- ........(((....(((((((((..(((((.(((.((.....--)).))).)))))((((-((((.....))))))))............)))))))))...))).- ( -23.70, z-score = -1.58, R) >droEre2.scaffold_4929 3422719 96 + 26641161 UUUGAGCUGUAAAUAAAAUAUAGAACCAUUUGGCACACUAACA--GCAUGUGAAAUGGAAA-UCGUGCCAAGCGGGGUCAGAAUCUGACUCUACGAGUA--------- ......(((((.......)))))..((((((.(((..(.....--)..))).))))))...-((((((...))((((((((...))))))))))))...--------- ( -23.00, z-score = -1.33, R) >droSim1.chr2L 3331519 96 + 22036055 UUUGAGCUGUAAAUAAAAUAUAUAACCAUUUGGCACACUAACA--GUAUGUGAAAUGGAAA-UCGUGCCAAGCGGGGUCAGAAUCUGACUUUAUGUGUG--------- .................((((((((((.((((((((....((.--....))((........-)))))))))).))((((((...)))))))))))))).--------- ( -23.10, z-score = -1.53, R) >droAna3.scaffold_12916 2855019 97 - 16180835 UUCCAGCUGUAAAUAAAAAACAUAACUAUUUGGCACACUAACA--GUCCAAAGAAUGGAAA-UCGUGCCAAGUCGGGUCAGCUGCUGGCACUGCCAGUGG-------- ..(((((((................(((((((((((.......--.((((.....))))..-..))))))))).))..)))))((((((...))))))))-------- ( -27.97, z-score = -1.93, R) >consensus UUUGAGCUGUAAAUAAAAUAUAUAACCAUUUGGCACACUAACA__GUAUGUGAAAUGGAAA_UCGUGCCAAGCGGGGUCAGAAUCUGACUCUAUUUGUA_________ .............................(((((((............................)))))))..................................... ( -8.17 = -7.90 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:13:46 2011